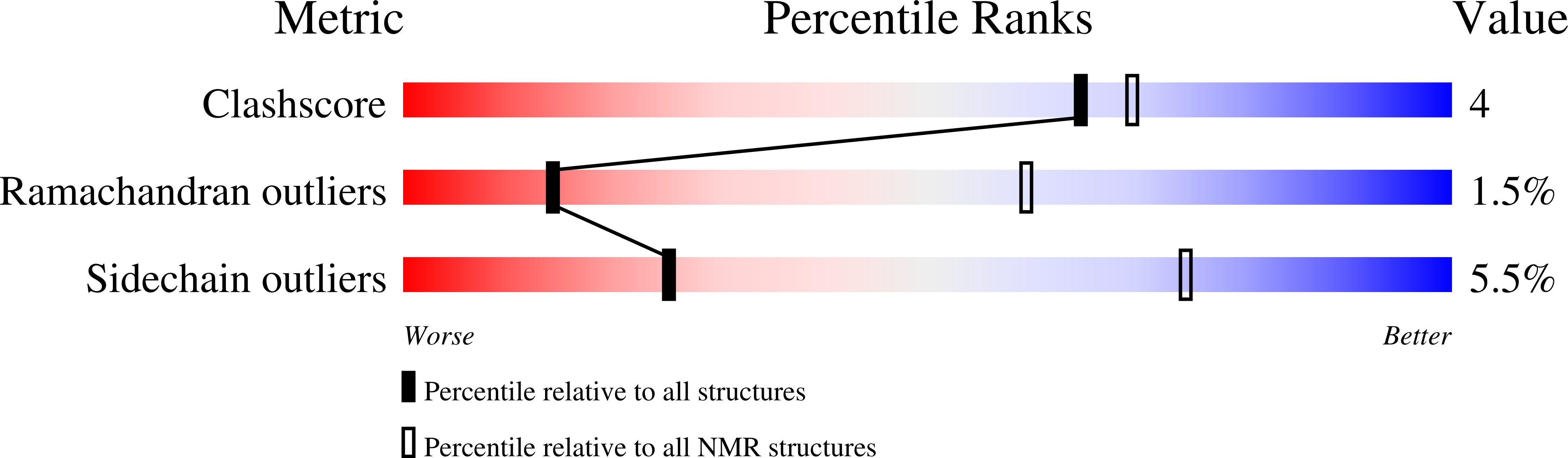A molecular explanation for the recessive nature of parkin-linked Parkinson's disease.
Spratt, D.E., Julio Martinez-Torres, R., Noh, Y.J., Mercier, P., Manczyk, N., Barber, K.R., Aguirre, J.D., Burchell, L., Purkiss, A., Walden, H., Shaw, G.S.(2013) Nat Commun 4: 1983-1983
- PubMed: 23770917
- DOI: https://doi.org/10.1038/ncomms2983
- Primary Citation of Related Structures:
2LWR, 2M48 - PubMed Abstract:
Mutations in the park2 gene, encoding the RING-inBetweenRING-RING E3 ubiquitin ligase parkin, cause 50% of autosomal recessive juvenile Parkinsonism cases. More than 70 known pathogenic mutations occur throughout parkin, many of which cluster in the inhibitory amino-terminal ubiquitin-like domain, and the carboxy-terminal RING2 domain that is indispensable for ubiquitin transfer. A structural rationale showing how autosomal recessive juvenile Parkinsonism mutations alter parkin function is still lacking. Here we show that the structure of parkin RING2 is distinct from canonical RING E3 ligases and lacks key elements required for E2-conjugating enzyme recruitment. Several pathogenic mutations in RING2 alter the environment of a single surface-exposed catalytic cysteine to inhibit ubiquitination. Native parkin adopts a globular inhibited conformation in solution facilitated by the association of the ubiquitin-like domain with the RING-inBetweenRING-RING C-terminus. Autosomal recessive juvenile Parkinsonism mutations disrupt this conformation. Finally, parkin autoubiquitinates only in cis, providing a molecular explanation for the recessive nature of autosomal recessive juvenile Parkinsonism.
Organizational Affiliation:
Department of Biochemistry, Schulich School of Medicine and Dentistry, University of Western Ontario, London, Ontario, Canada N6A 5C1.