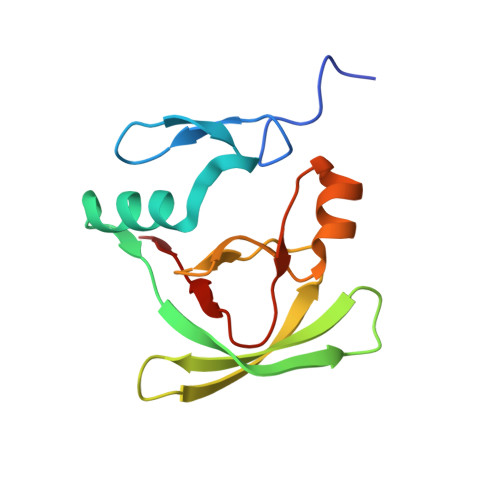
Protein-Protein Interactions as a Strategy towards Protein-Specific Drug Design: The Example of Ataxin-1.
de Chiara, C., Menon, R.P., Kelly, G., Pastore, A.(2013) PLoS One 8: e76456-e76456
- PubMed: 24155902
- DOI: https://doi.org/10.1371/journal.pone.0076456
- Primary Citation of Related Structures:
2M41 - PubMed Abstract:
A main challenge for structural biologists is to understand the mechanisms that discriminate between molecular interactions and determine function. Here, we show how partner recognition of the AXH domain of the transcriptional co-regulator ataxin-1 is fine-tuned by a subtle balance between self- and hetero-associations. Ataxin-1 is the protein responsible for the hereditary spinocerebellar ataxia type 1, a disease linked to protein aggregation and transcriptional dysregulation. Expansion of a polyglutamine tract is essential for ataxin-1 aggregation, but the sequence-wise distant AXH domain plays an important aggravating role in the process. The AXH domain is also a key element for non-aberrant function as it intervenes in interactions with multiple protein partners. Previous data have shown that AXH is dimeric in solution and forms a dimer of dimers when crystallized. By solving the structure of a complex of AXH with a peptide from the interacting transcriptional repressor CIC, we show that the dimer interface of AXH is displaced by the new interaction and that, when blocked by the CIC peptide AXH aggregation and misfolding are impaired. This is a unique example in which palindromic self- and hetero-interactions within a sequence with chameleon properties discriminate the partner. We propose a drug design strategy for the treatment of SCA1 that is based on the information gained from the AXH/CIC complex.
Organizational Affiliation:
National Institute for Medical Research of the Medical Research Council, London, United Kingdom.