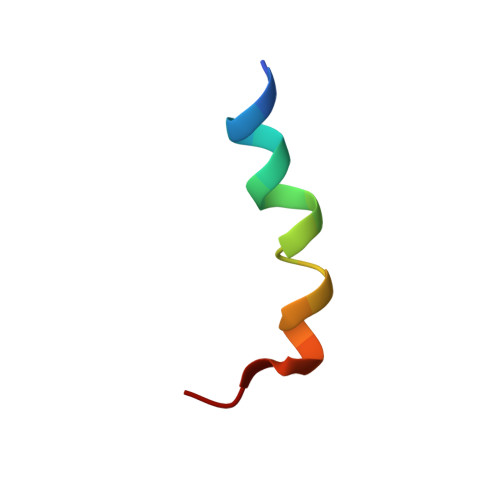
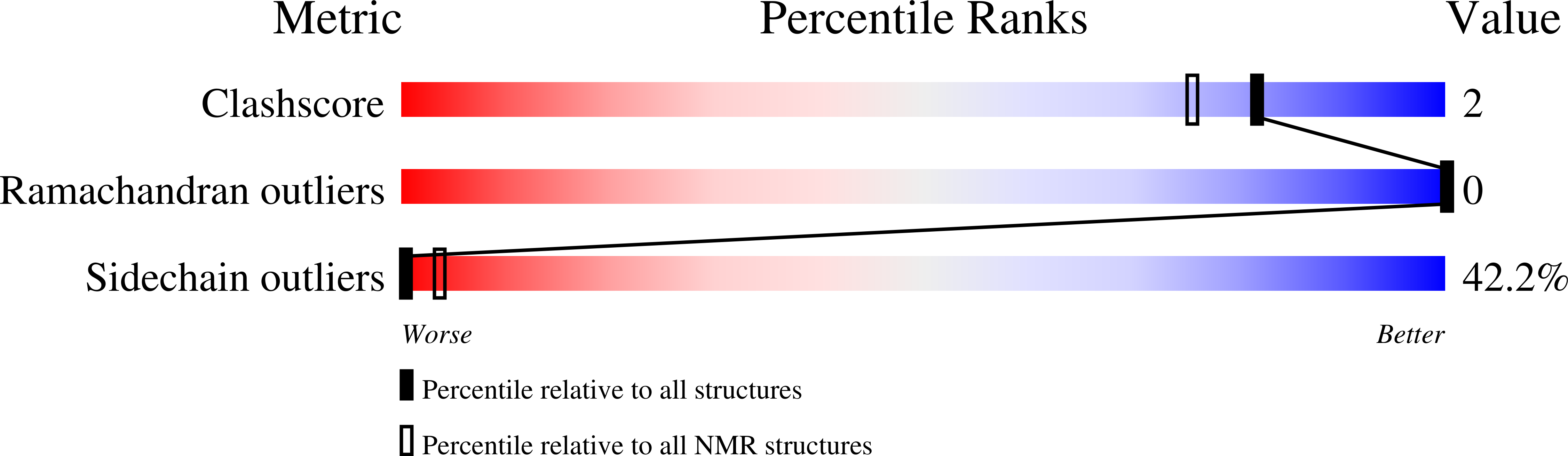Structure and Orientation of the gH625-644 Membrane Interacting Region of Herpes Simplex Virus Type 1 in a Membrane Mimetic System.
Galdiero, S., Russo, L., Falanga, A., Cantisani, M., Vitiello, M., Fattorusso, R., Malgieri, G., Galdiero, M., Isernia, C.(2012) Biochemistry 51: 3121-3128
- PubMed: 22397737
- DOI: https://doi.org/10.1021/bi201589m
- Primary Citation of Related Structures:
2LQY - PubMed Abstract:
Glycoprotein H (gH) of the herpes simplex virus type 1 is involved in the complex mechanism of membrane fusion of the viral envelope with host cells. The virus requires four glycoproteins (gB, gD, gH, gL) to execute fusion and the role played by gH remains mysterious. Mutational studies have revealed several regions of gH ectodomain required for fusion and identified the segment from amino acid 625 to 644 as the most fusogenic region. Here, we studied the behavior in a membrane-mimicking DPC micellar environment of a peptide encompassing this region (gH625-644) and determined its NMR solution structure and its orientation within the micelles.
Organizational Affiliation:
Department of Biological Sciences, Division of Biostructures, University of Naples Federico II, Napoli, Italy.