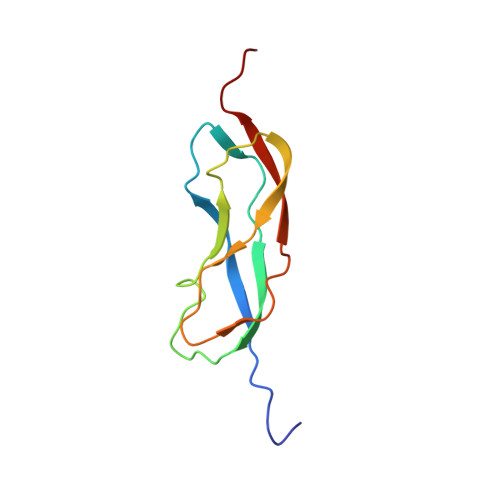
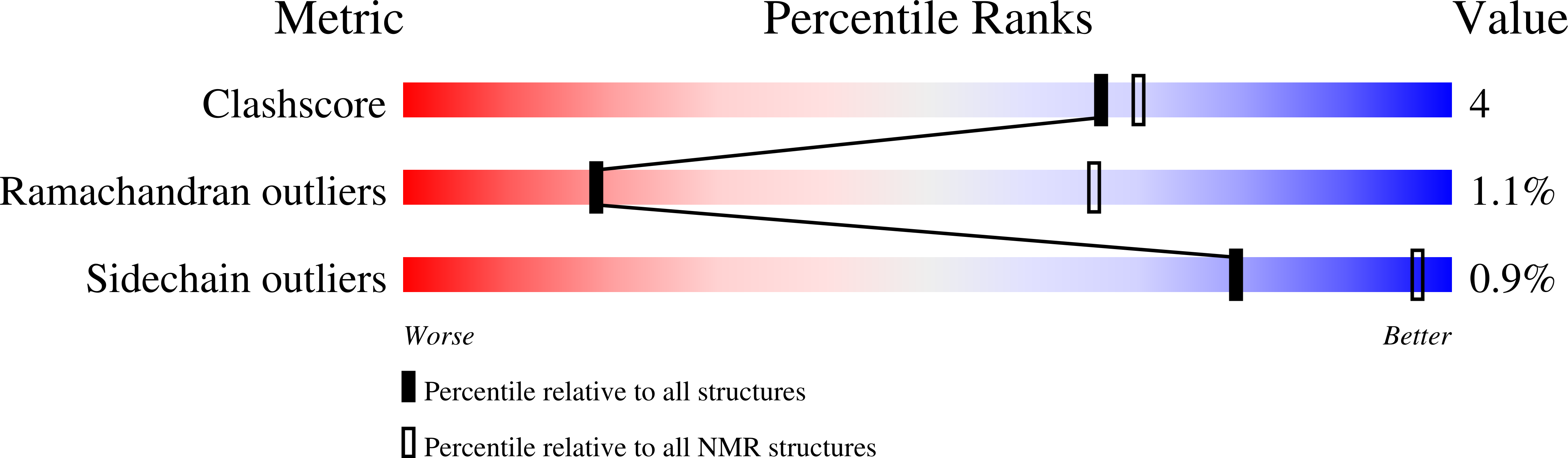Structures of domains I and IV from YbbR are representative of a widely distributed protein family.
Barb, A.W., Cort, J.R., Seetharaman, J., Lew, S., Lee, H.W., Acton, T., Xiao, R., Kennedy, M.A., Tong, L., Montelione, G.T., Prestegard, J.H.(2011) Protein Sci 20: 396-405
- PubMed: 21154411
- DOI: https://doi.org/10.1002/pro.571
- Primary Citation of Related Structures:
2KPU, 2L3U, 2L5N - PubMed Abstract:
YbbR domains are widespread throughout Eubacteria and are expressed as monomeric units, linked in tandem repeats or cotranslated with other domains. Although the precise role of these domains remains undefined, the location of the multiple YbbR domain-encoding ybbR gene in the Bacillus subtilis glmM operon and its previous identification as a substrate for a surfactin-type phosphopantetheinyl transferase suggests a role in cell growth, division, and virulence. To further characterize the YbbR domains, structures of two of the four domains (I and IV) from the YbbR-like protein of Desulfitobacterium hafniense Y51 were solved by solution nuclear magnetic resonance and X-ray crystallography. The structures show the domains to have nearly identical topologies despite a low amino acid identity (23%). The topology is dominated by β-strands, roughly following a "figure 8" pattern with some strands coiling around the domain perimeter and others crossing the center. A similar topology is found in the C-terminal domain of two stress-responsive bacterial ribosomal proteins, TL5 and L25. Based on these models, a structurally guided amino acid alignment identifies features of the YbbR domains that are not evident from naïve amino acid sequence alignments. A structurally conserved cis-proline (cis-Pro) residue was identified in both domains, though the local structure in the immediate vicinities surrounding this residue differed between the two models. The conservation and location of this cis-Pro, plus anchoring Val residues, suggest this motif may be significant to protein function.
Organizational Affiliation:
Complex Carbohydrate Research Center, The University of Georgia, Athens, Georgia, USA.