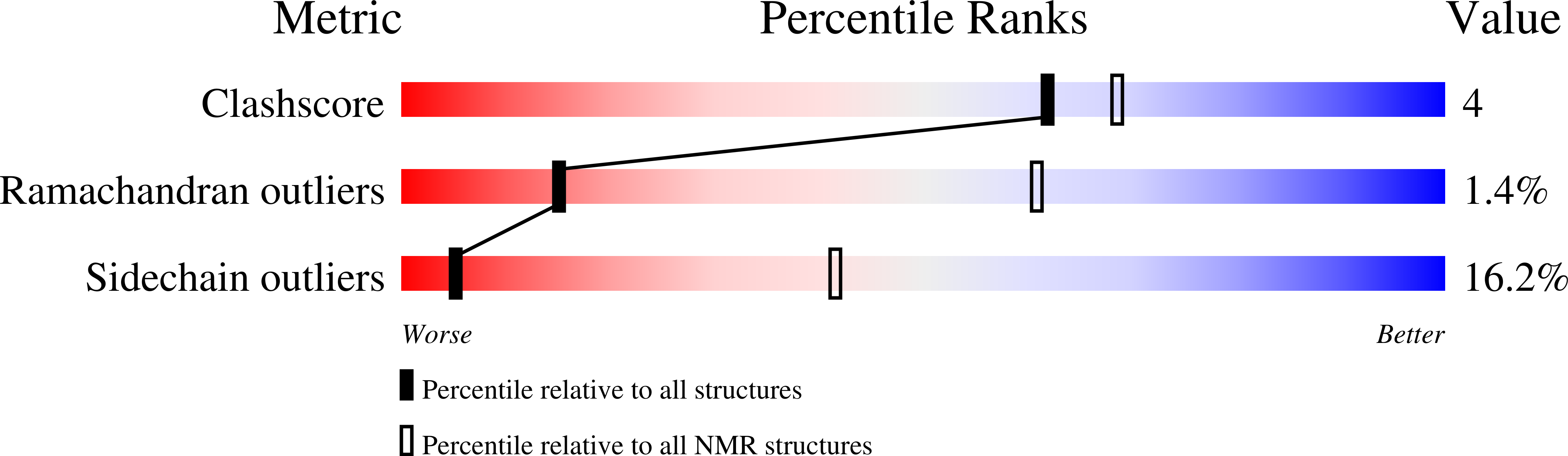The copper-responsive repressor CopR of Lactococcus lactis is a 'winged helix' protein.
Cantini, F., Banci, L., Solioz, M.(2009) Biochem J 417: 493-499
- PubMed: 18837698
- DOI: https://doi.org/10.1042/BJ20081713
- Primary Citation of Related Structures:
2K4B - PubMed Abstract:
CopR of Lactococcus lactis is a copper-responsive repressor involved in copper homoeostasis. It controls the expression of a total of 11 genes, the CopR regulon, in a copper-dependent manner. In the absence of copper, CopR binds to the promoters of the CopR regulon. Copper releases CopR from the promoters, allowing transcription of the downstream genes to proceed. CopR binds through its N-terminal domain to a 'cop box' of consensus TACANNTGTA, which is conserved in Firmicutes. We have solved the NMR solution structure of the N-terminal DNA-binding domain of CopR. The protein fold has a winged helix structure resembling that of the BlaI repressor which regulates antibiotic resistance in Bacillus licheniformis. CopR differs from other copper-responsive repressors, and the present structure represents a novel family of copper regulators, which we propose to call the CopY family.
Organizational Affiliation:
Department of Chemistry and Magnetic Resonance Center, University of Florence, Sesto Fiorentino, Italy.