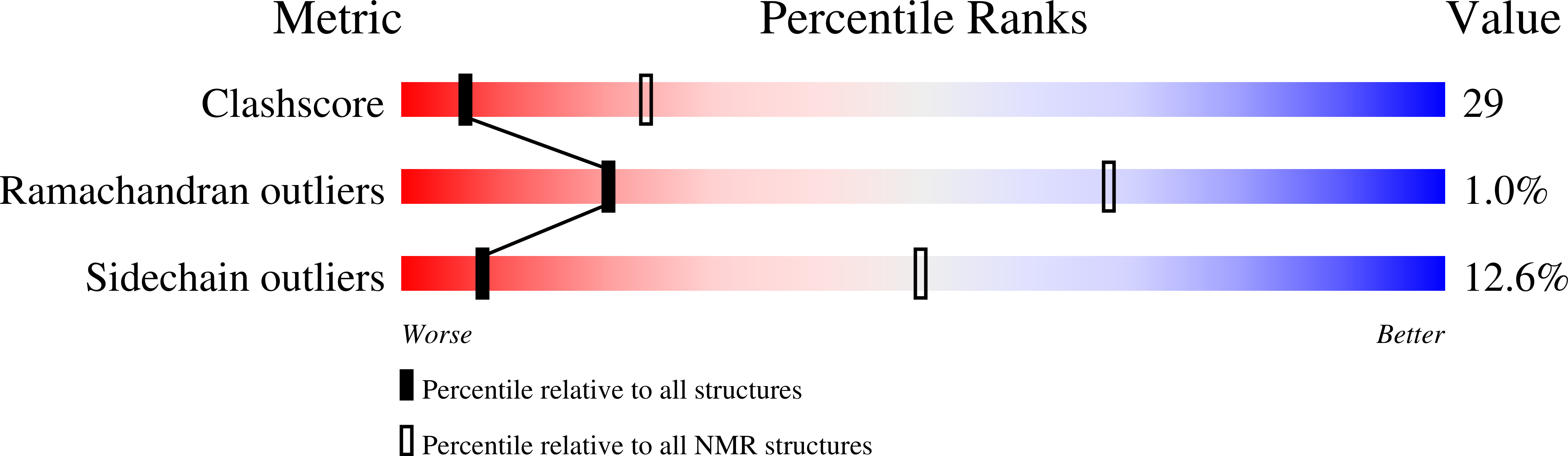Solution structure and phylogenetics of Prod1, a member of the three-finger protein superfamily implicated in salamander limb regeneration.
Garza-Garcia, A., Harris, R., Esposito, D., Gates, P.B., Driscoll, P.C.(2009) PLoS One 4: e7123-e7123
- PubMed: 19771161
- DOI: https://doi.org/10.1371/journal.pone.0007123
- Primary Citation of Related Structures:
2JVE - PubMed Abstract:
Following the amputation of a limb, newts and salamanders have the capability to regenerate the lost tissues via a complex process that takes place at the site of injury. Initially these cells undergo dedifferentiation to a state competent to regenerate the missing limb structures. Crucially, dedifferentiated cells have memory of their level of origin along the proximodistal (PD) axis of the limb, a property known as positional identity. Notophthalmus viridescens Prod1 is a cell-surface molecule of the three-finger protein (TFP) superfamily involved in the specification of newt limb PD identity. The TFP superfamily is a highly diverse group of metazoan proteins that includes snake venom toxins, mammalian transmembrane receptors and miscellaneous signaling molecules. With the aim of identifying potential orthologs of Prod1, we have solved its 3D structure and compared it to other known TFPs using phylogenetic techniques. The analysis shows that TFP 3D structures group in different categories according to function. Prod1 clusters with other cell surface protein TFP domains including the complement regulator CD59 and the C-terminal domain of urokinase-type plasminogen activator. To infer orthology, a structure-based multiple sequence alignment of representative TFP family members was built and analyzed by phylogenetic methods. Prod1 has been proposed to be the salamander CD59 but our analysis fails to support this association. Prod1 is not a good match for any of the TFP families present in mammals and this result was further supported by the identification of the putative orthologs of both CD59 and N. viridescens Prod1 in sequence data for the salamander Ambystoma tigrinum. The available data suggest that Prod1, and thereby its role in encoding PD identity, is restricted to salamanders. The lack of comparable limb-regenerative capability in other adult vertebrates could be correlated with the absence of the Prod1 gene.
Organizational Affiliation:
Division of Molecular Structure, MRC National Institute for Medical Research, London, United Kingdom.