Structural insights into activation of phosphatidylinositol 4-kinase (Pik1) by yeast frequenin (Frq1).
Strahl, T., Huttner, I.G., Lusin, J.D., Osawa, M., King, D., Thorner, J., Ames, J.B.(2007) J Biol Chem 282: 30949-30959
- PubMed: 17720810
- DOI: https://doi.org/10.1074/jbc.M705499200
- Primary Citation of Related Structures:
2JU0 - PubMed Abstract:
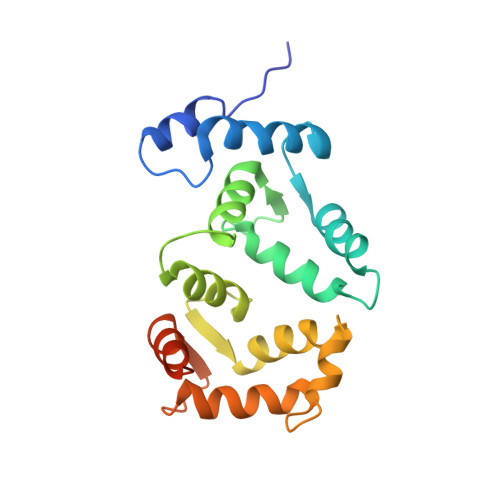
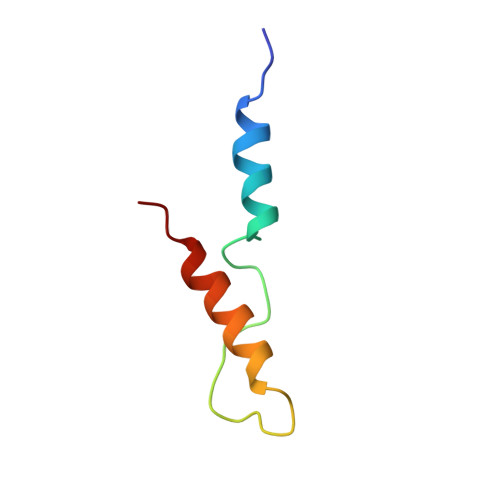
Yeast frequenin (Frq1), a small N-myristoylated EF-hand protein, activates phosphatidylinositol 4-kinase Pik1. The NMR structure of Ca2+-bound Frq1 complexed to an N-terminal Pik1 fragment (residues 121-174) was determined. The Frq1 main chain is similar to that in free Frq1 and related proteins in the same branch of the calmodulin superfamily. The myristoyl group and first eight residues of Frq1 are solvent-exposed, and Ca2+ binds the second, third, and fourth EF-hands, which associate to create a groove with two pockets. The Pik1 peptide forms two helices (125-135 and 156-169) connected by a 20-residue loop. Side chains in the Pik1 N-terminal helix (Val-127, Ala-128, Val-131, Leu-132, and Leu-135) interact with solvent-exposed residues in the Frq1 C-terminal pocket (Leu-101, Trp-103, Val-125, Leu-138, Ile-152, and Leu-155); side chains in the Pik1 C-terminal helix (Ala-157, Ala-159, Leu-160, Val-161, Met-165, and Met-167) contact solvent-exposed residues in the Frq1 N-terminal pocket (Trp-30, Phe-34, Phe-48, Ile-51, Tyr-52, Phe-55, Phe-85, and Leu-89). This defined complex confirms that residues in Pik1 pinpointed as necessary for Frq1 binding by site-directed mutagenesis are indeed sufficient for binding. Removal of the Pik1 N-terminal region (residues 8-760) from its catalytic domain (residues 792-1066) abolishes lipid kinase activity, inconsistent with Frq1 binding simply relieving an autoinhibitory constraint. Deletion of the lipid kinase unique motif (residues 35-110) also eliminates Pik1 activity. In the complex, binding of Ca2+-bound Frq1 forces the Pik1 chain into a U-turn. Frq1 may activate Pik1 by facilitating membrane targeting via the exposed N-myristoyl group and by imposing a structural transition that promotes association of the lipid kinase unique motif with the kinase domain.
Organizational Affiliation:
Department of Molecular and Cell Biology, Division of Biochemistry and Molecular Biology, University of California, Berkeley, California 94720-3202, USA.