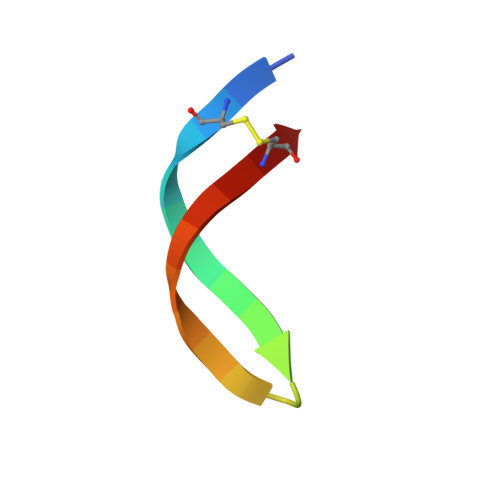
Structure and mode of action of the antimicrobial peptide arenicin
Andra, J., Jakovkin, I., Grotzinger, J., Hecht, O., Krasnosdembskaya, A.D., Goldmann, T., Gutsmann, T., Leippe, M.(2008) Biochem J 410: 113-122
- PubMed: 17935487
- DOI: https://doi.org/10.1042/BJ20071051
- Primary Citation of Related Structures:
2JSB - PubMed Abstract:
The solution structure and the mode of action of arenicin isoform 1, an antimicrobial peptide with a unique 18-residue loop structure, from the lugworm Arenicola marina were elucidated here. Arenicin folds into a two-stranded antiparallel beta-sheet. It exhibits high antibacterial activity at 37 and 4 degrees C against Gram-negative bacteria, including polymyxin B-resistant Proteus mirabilis. Bacterial killing occurs within minutes and is accompanied by membrane permeabilization, membrane detachment and release of cytoplasm. Interaction of arenicin with reconstituted membranes that mimic the lipopolysaccharide-containing outer membrane or the phospholipid-containing plasma membrane of Gram-negative bacteria exhibited no pronounced lipid specificity. Arenicin-induced current fluctuations in planar lipid bilayers correspond to the formation of short-lived heterogeneously structured lesions. Our results strongly suggest that membrane interaction plays a pivotal role in the antibacterial activity of arenicin.
Organizational Affiliation:
Research Center Borstel, Leibniz-Center for Medicine and Biosciences, Parkallee 10, 23845 Borstel, Germany. jandrae@fz-borstel.de