Synthesis, solution structure and immune recognition of an epidermal growth factor-like domain from Plasmodium falciparum merozoite surface protein-1.
James, S., Moehle, K., Renard, A., Mueller, M.S., Vogel, D., Zurbriggen, R., Pluschke, G., Robinson, J.A.(2006) Chembiochem 7: 1943-1950
- PubMed: 17068840
- DOI: https://doi.org/10.1002/cbic.200600357
- Primary Citation of Related Structures:
2FLG - PubMed Abstract:
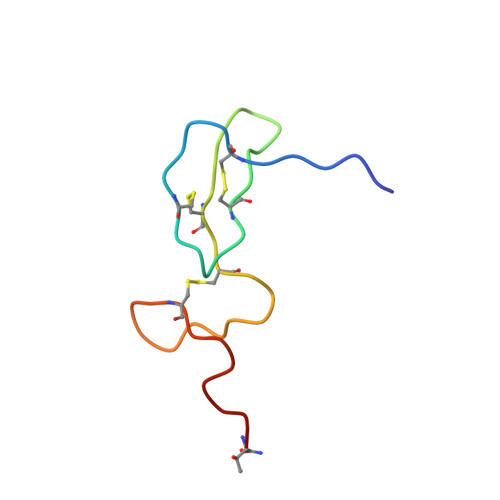
The Plasmodium falciparum merozoite surface protein-1 19 kDa fragment (MSP-1(19)) comprises two closely packed EGF-like domains (EGF=epidermal growth factor), each stabilized by three disulfide bonds. The native conformation of this protein is important for eliciting P. falciparum growth inhibitory antibodies. Here we show that the N-terminal EGF domain alone can be chemically synthesized and efficiently refolded to a native-like state, as shown by its solution structure as determined by NMR spectroscopy. In order to study its immunogenicity, the domain was coupled through its N terminus to a phospholipid and incorporated into reconstituted influenza virus-like particles (virosomes). When used to immunize mice, the peptide-loaded virosomes elicited potent humoral immune responses that were shown by Western blots and immunofluorescence assays to cross-react with native MSP-1 on the surfaces of P. falciparum blood stage parasites. This opens the way for a medicinal chemistry-oriented approach to the study and optimization of the antigenicity of the protein as a potential malaria vaccine candidate, whilst exploiting the immunopotentiating properties of influenza virosomes as a delivery vehicle.
Organizational Affiliation:
Department of Chemistry, University of Zürich, Winterthurerstrasse 190, 8057 Zürich, Switzerland.