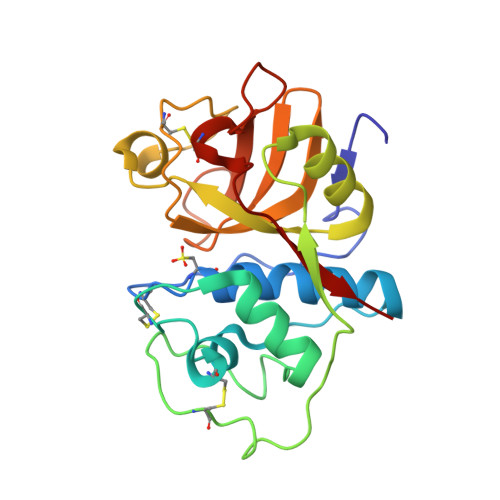
High-Resolution Complex of Papain with Remnants of a Cysteine Protease Inhibitor Derived from Trypanosoma Brucei
Alphey, M.S., Hunter, W.N.(2006) Acta Crystallogr Sect F Struct Biol Cryst Commun 62: 504
- PubMed: 16754967
- DOI: https://doi.org/10.1107/S1744309106014849
- Primary Citation of Related Structures:
2CIO - PubMed Abstract:
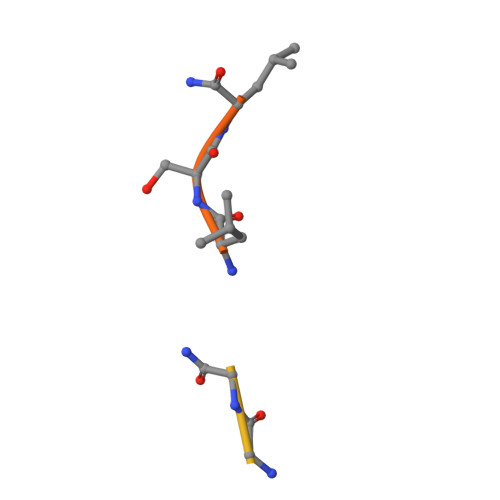
Attempts to cocrystallize the cysteine protease papain derived from the latex of Carica papaya with an inhibitor of cysteine proteases (ICP) from Trypanosoma brucei were unsuccessful. However, crystals of papain that diffracted to higher resolution, 1.5 A, than other crystals of this archetypal cysteine protease were obtained, so the analysis was continued. Surprisingly, the substrate-binding cleft was occupied by two short peptide fragments which have been assigned as remnants of ICP. Comparisons reveal that these peptides bind in the active site in a manner similar to that of the human cysteine protease inhibitor stefin B when it is complexed to papain. The assignment of the fragment sequences is consistent with the specificity of the protease.
- Division of Biological Chemistry and Molecular Microbiology, School of Life Sciences, University of Dundee, Dundee DD1 5EH, Scotland.
Organizational Affiliation: