Structure of ESCRT-II endosomal trafficking complex
Hierro, A., Sun, J., Rusnak, A.S., Kim, J., Prag, G., Emr, S.D., Hurley, J.H.(2004) Nature 431: 221-225
- PubMed: 15329733
- DOI: https://doi.org/10.1038/nature02914
- Primary Citation of Related Structures:
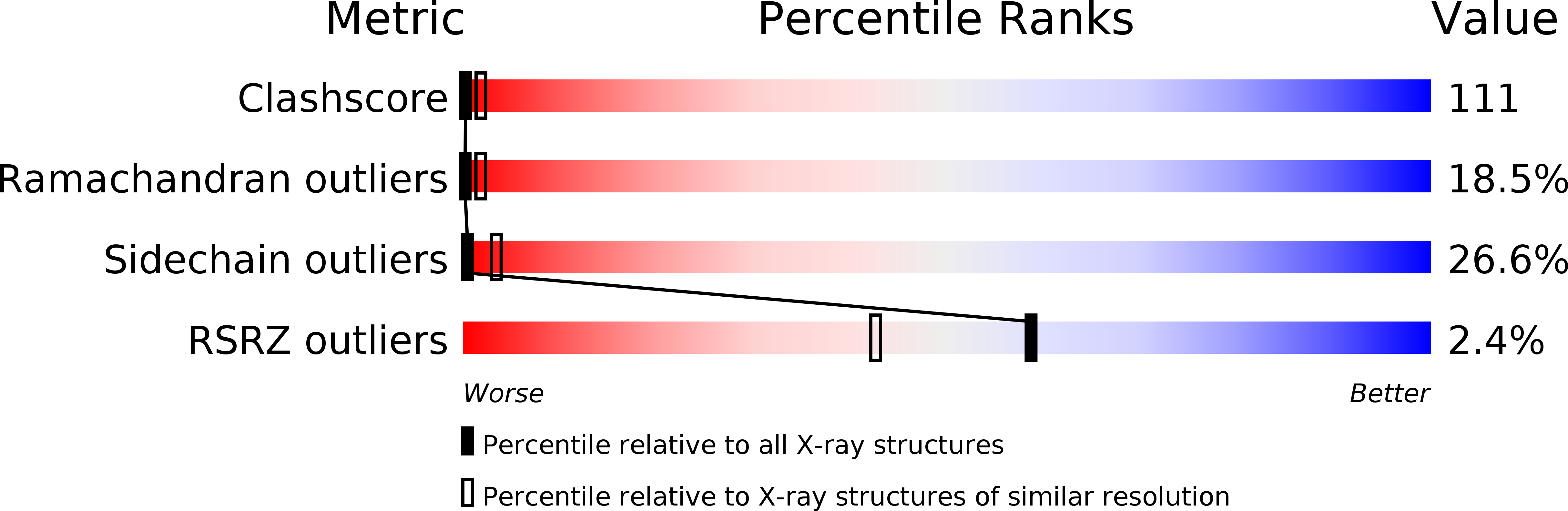
1U5T - PubMed Abstract:
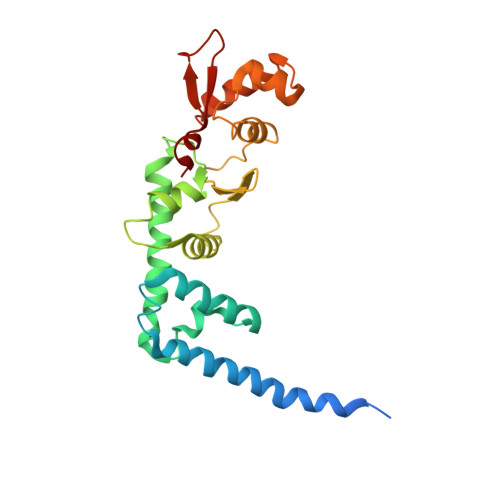
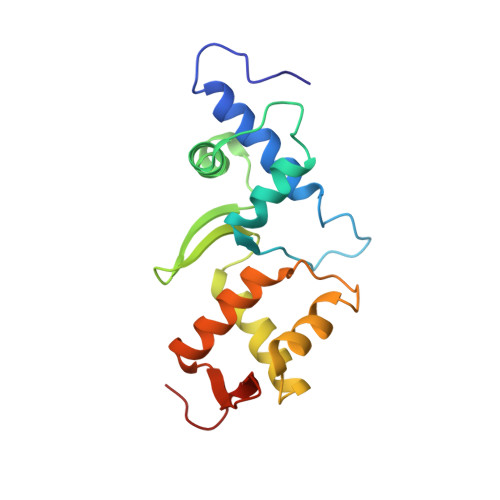
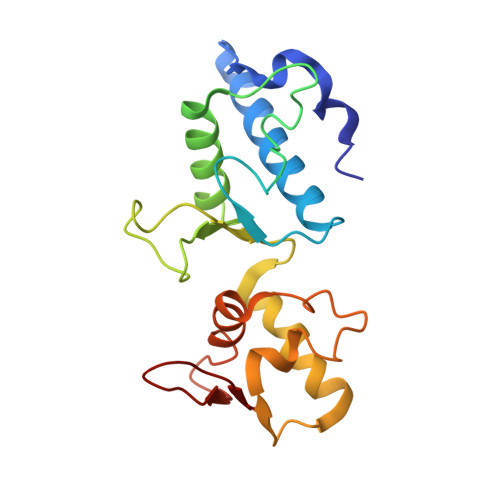
The multivesicular-body (MVB) pathway delivers transmembrane proteins and lipids to the lumen of the endosome. The multivesicular-body sorting pathway has crucial roles in growth-factor-receptor downregulation, developmental signalling, regulation of the immune response and the budding of certain enveloped viruses such as human immunodeficiency virus. Ubiquitination is a signal for sorting into the MVB pathway, which also requires the functions of three protein complexes, termed ESCRT-I, -II and -III (endosomal sorting complex required for transport). Here we report the crystal structure of the core of the yeast ESCRT-II complex, which contains one molecule of the Vps protein Vps22, the carboxy-terminal domain of Vps36 and two molecules of Vps25, and has the shape of a capital letter 'Y'. The amino-terminal coiled coil of Vps22 and the flexible linker leading to the ubiquitin-binding NZF domain of Vps36 both protrude from the tip of one branch of the 'Y'. Vps22 and Vps36 form nearly equivalent interactions with the two Vps25 molecules at the centre of the 'Y'. The structure suggests how ubiquitinated cargo could be passed between ESCRT components of the MVB pathway through the sequential transfer of ubiquitinated cargo from one complex to the next.
Organizational Affiliation:
Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, US Department of Health and Human Services, Bethesda, Maryland 20892-0580, USA.