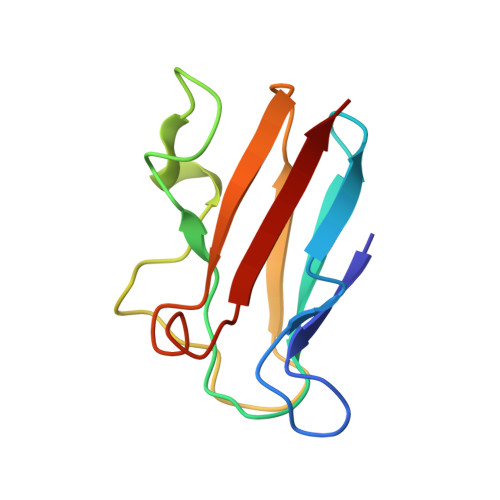
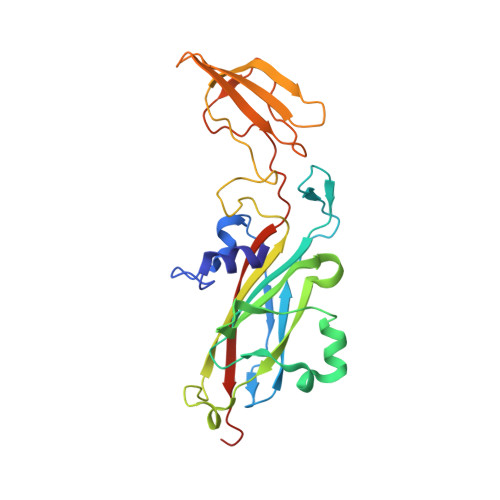
The transient complex of poplar plastocyanin with cytochrome f: effects of ionic strength and pH
Lange, C., Cornvik, T., Diaz-Moreno, I., Ubbink, M.(2005) Biochim Biophys Acta 1707: 179-188
- PubMed: 15863096
- DOI: https://doi.org/10.1016/j.bbabio.2004.12.002
- Primary Citation of Related Structures:
1TKW - PubMed Abstract:
The orientation of poplar plastocyanin in the complex with turnip cytochrome f has been determined by rigid-body calculations using restraints from paramagnetic NMR measurements. The results show that poplar plastocyanin interacts with cytochrome f with the hydrophobic patch of plastocyanin close to the heme region on cytochrome f and via electrostatic interactions between the charged patches on both proteins. Plastocyanin is tilted relative to the orientation reported for spinach plastocyanin, resulting in a longer distance between iron and copper (13.9 A). With increasing ionic strength, from 0.01 to 0.11 M, all observed chemical-shift changes decrease uniformly, supporting the idea that electrostatic forces contribute to complex formation. There is no indication for a rearrangement of the transient complex in this ionic strength range, contrary to what had been proposed earlier on the basis of kinetic data. By decreasing the pH from pH 7.7 to pH 5.5, the complex is destabilized. This may be attributed to the protonation of the conserved acidic patches or the copper ligand His87 in poplar plastocyanin, which are shown to have similar pK(a) values. The results are interpreted in a two-step model for complex formation.
Organizational Affiliation:
Leiden Institute of Chemistry, Leiden University, Gorlaeus Laboratories, P.O. Box 9502, 2300 RA Leiden, The Netherlands.