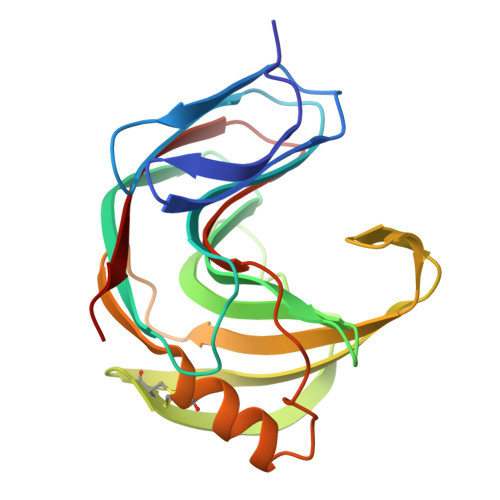
The tertiary structure at 1.59 A resolution and the proposed amino acid sequence of a family-11 xylanase from the thermophilic fungus Paecilomyces varioti bainier.
Kumar, P.R., Eswaramoorthy, S., Vithayathil, P.J., Viswamitra, M.A.(2000) J Mol Biol 295: 581-593
- PubMed: 10623548
- DOI: https://doi.org/10.1006/jmbi.1999.3348
- Primary Citation of Related Structures:
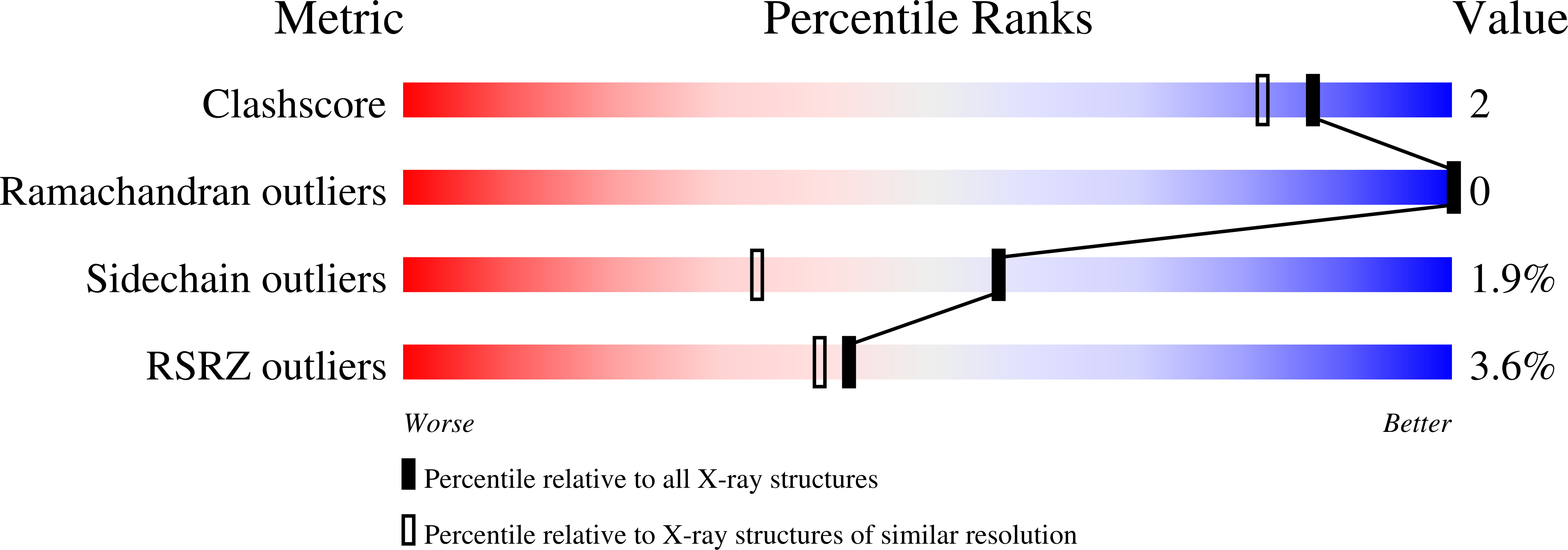
1PVX - PubMed Abstract:
We report the crystal structure at 1.59 A and the proposed amino acid sequence of an endo-1,4-beta-xylanase (PVX) from the thermophilic fungus Paecilomyces varioti Bainier (PvB), stable up to 75 degrees C. This fungus is attracting clinical attention as a pathogen causing post-surgical infections. Its xylanase, known as a skin-contact allergen, is the first protein from this fungus whose three-dimensional structure has been elucidated. The crystals of PVX conform to the space group P2(1)2(1)2(1 )with a=38.76 A, b=54.06 A and c=90.06 A. The structure was solved by molecular replacement techniques using polyalanine coordinates of the Thermomyces lanuginosus xylanase (PDB code 1YNA) and a careful model building based on the amino acid sequence known for two trypsin-digested peptide fragments (17 residues), the sequence and structural alignment of family-11 xylanases and electron density maps. The final refined model has 194 amino acid residues and 128 water molecules, with a crystallographic R-factor of 19.07 % and a free R-factor of 21.94 %. The structure belongs to an all-beta fold, with two curved beta-sheets, forming the cylindrical active-site cleft, and a lone alpha-helix, as present in other family-11 xylanases. We have carried out a quantitative comparison of the structure and sequence of the present thermophilic xylanase (PVX) with other available native structures of mesophiles and thermophiles, the first such detailed analysis to be carried out on family-11 xylanases. The analysis provides a basis for the rationalisation of the idea that the "hinge" region is made more compact in thermophiles by the addition of a disulphide bridge between Cys110 and Cys154 and a N-H.O hydrogen bond between Trp159 near the extremity of the lone alpha-helix and Trp138 on beta-strand B8. This work brings out explicitly the presence of the C-H.O and the C-H.pi type interactions in these enzymes. A complete description of structural stability of these enzymes needs to take account of these weaker interactions.
Organizational Affiliation:
Department of Physics, Indian Institute of Science, Bangalore, 560 012, India.