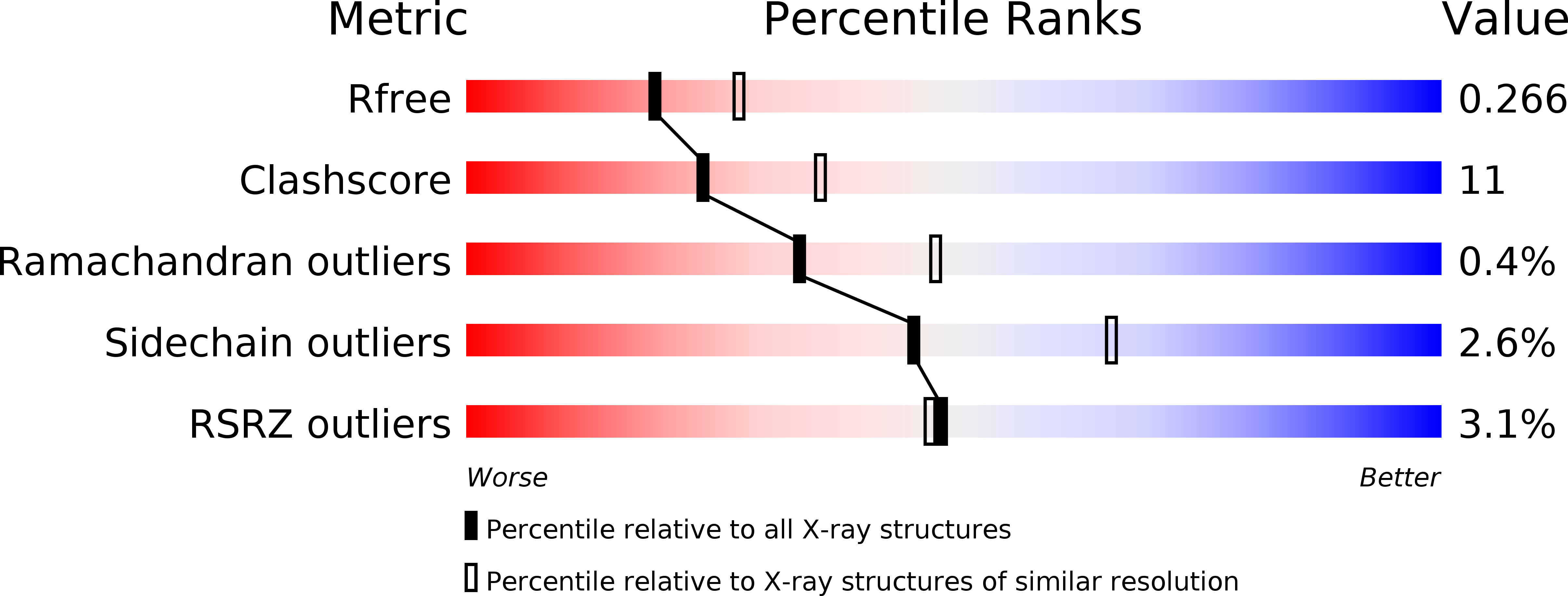
Self-association of the H3 region of syntaxin 1A. Implications for intermediates in SNARE complex assembly.
Misura, K.M., Scheller, R.H., Weis, W.I.(2001) J Biol Chem 276: 13273-13282
- PubMed: 11118447
- DOI: https://doi.org/10.1074/jbc.M009636200
- Primary Citation of Related Structures:
1HVV - PubMed Abstract:
Intracellular membrane fusion requires SNARE proteins found on the vesicle and target membranes. SNAREs associate by formation of a parallel four-helix bundle, and it has been suggested that formation of this complex promotes membrane fusion. The membrane proximal region of the cytoplasmic domain of the SNARE syntaxin 1A, designated H3, contributes one of the four helices to the SNARE complex. In the crystal structure of syntaxin 1A H3, four molecules associate as a homotetramer composed of two pairs of parallel helices that are anti-parallel to each other. The H3 oligomer observed in the crystals is also found in solution, as assessed by gel filtration and chemical cross-linking studies. The crystal structure reveals that the highly conserved Phe-216 packs against conserved Gln-226 residues present on the anti-parallel pair of helices. Modeling indicates that Phe-216 prevents parallel tetramer formation. Mutation of Phe-216 to Leu appears to allow formation of parallel tetramers, whereas mutation to Ala destabilizes the protein. These results indicate that Phe-216 has a role in preventing formation of stable parallel helical bundles, thus favoring the interaction of the H3 region of syntaxin 1a with other proteins involved in membrane fusion.
Organizational Affiliation:
Department of Structural Biology, The Howard Hughes Medical Institute, Stanford University School of Medicine, Stanford, California 94305, USA.