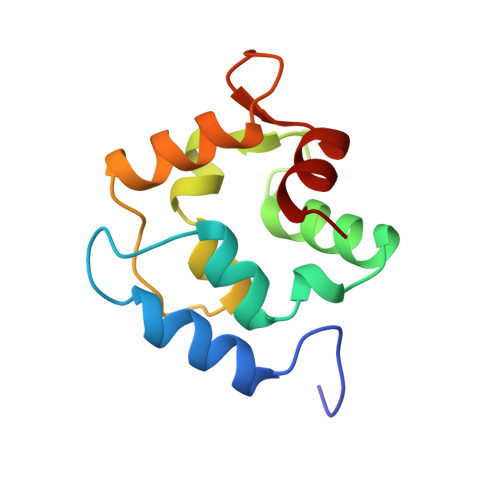
X-Ray crystal structure and molecular dynamics simulations of silver hake parvalbumin (Isoform B).
Richardson, R.C., King, N.M., Harrington, D.J., Sun, H., Royer, W.E., Nelson, D.J.(2000) Protein Sci 9: 73-82
- PubMed: 10739249
- DOI: https://doi.org/10.1110/ps.9.1.73
- Primary Citation of Related Structures:
1BU3 - PubMed Abstract:
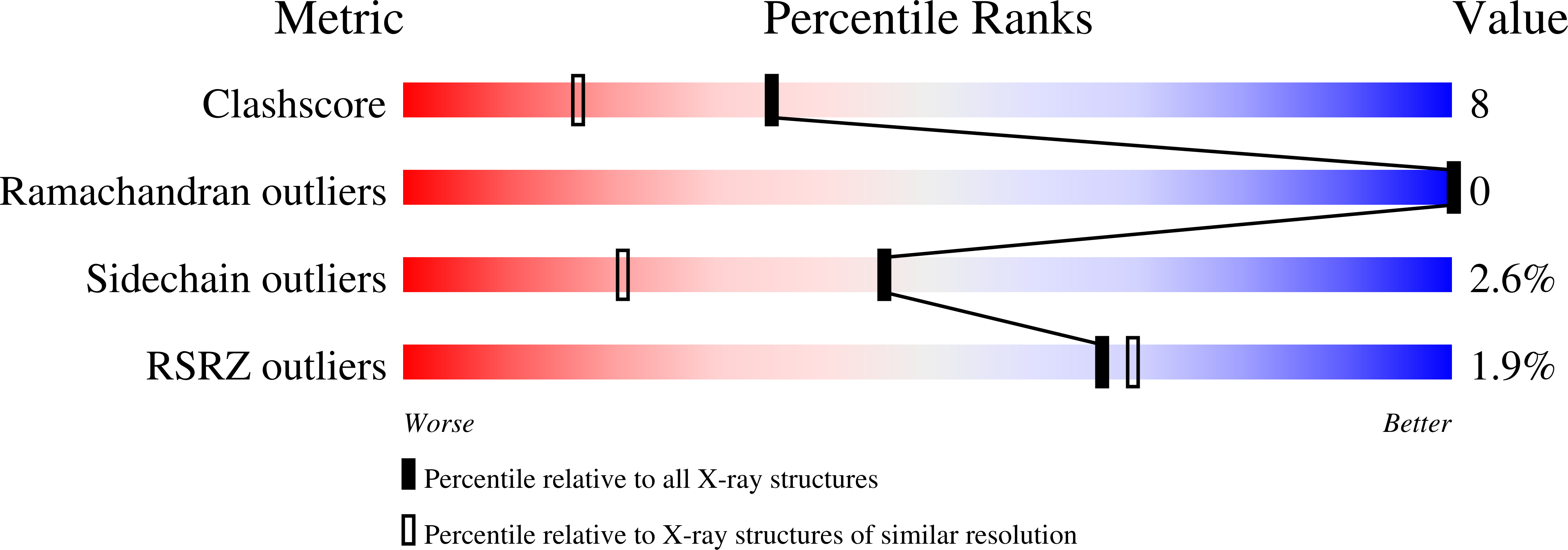
Parvalbumins constitute a class of calcium-binding proteins characterized by the presence of several helix-loop-helix (EF-hand) motifs. In a previous study (Revett SP, King G, Shabanowitz J, Hunt DF, Hartman KL, Laue TM, Nelson DJ, 1997, Protein Sci 7:2397-2408), we presented the sequence of the major parvalbumin isoform from the silver hake (Merluccius bilinearis) and presented spectroscopic and structural information on the excised "EF-hand" portion of the protein. In this study, the X-ray crystal structure of the silver hake major parvalbumin has been determined to high resolution, in the frozen state, using the molecular replacement method with the carp parvalbumin structure as a starting model. The crystals are orthorhombic, space group C2221, with a = 75.7 A, b = 80.7 A, and c = 42.1 A. Data were collected from a single crystal grown in 15% glycerol, which served as a cryoprotectant for flash freezing at -188 degrees C. The structure refined to a conventional R-value of 21% (free R 25%) for observed reflections in the range 8 to 1.65 A [1 > 2sigma(I)]. The refined model includes an acetylated amino terminus, 108 residues (characteristic of a beta parvalbumin lineage), 2 calcium ions, and 114 water molecules per protein molecule. The resulting structure was used in molecular dynamics (MD) simulations focused primarily on the dynamics of the ligands coordinating the Ca2+ ions in the CD and EF sites. MD simulations were performed on both the fully Ca2+ loaded protein and on a Ca2+ deficient variant, with Ca2+ only in the CD site. There was substantial agreement between the MD and X-ray results in addressing the issue of mobility of key residues in the calcium-binding sites, especially with regard to the side chain of Ser55 in the CD site and Asp92 in the EF site.
Organizational Affiliation:
Gustaf H. Carlson School of Chemistry, Clark University, Worcester, Massachusetts 01610, USA.