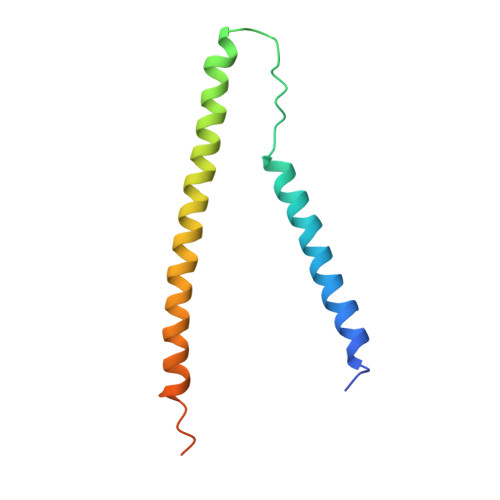
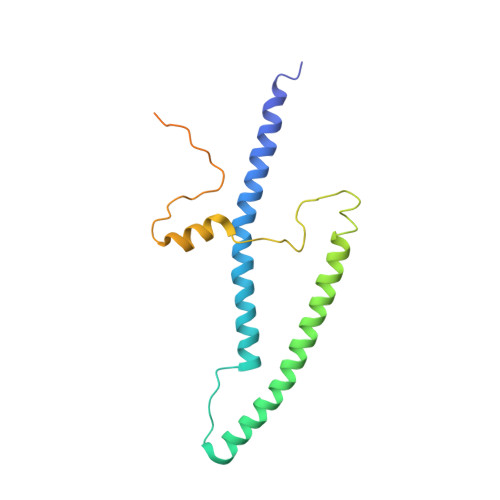
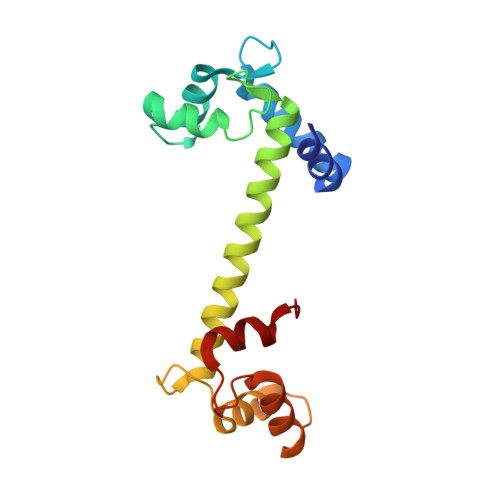
Ca2+-regulated structural changes in troponin
Vinogradova, M.V., Stone, D.B., Malanina, G.G., Karatzaferi, C., Cooke, R., Mendelson, R.A., Fletterick, R.J.(2005) Proc Natl Acad Sci U S A 102: 5038-5043
- PubMed: 15784741
- DOI: https://doi.org/10.1073/pnas.0408882102
- Primary Citation of Related Structures:
1YTZ, 1YV0 - PubMed Abstract:
Troponin senses Ca2+ to regulate contraction in striated muscle. Structures of skeletal muscle troponin composed of TnC (the sensor), TnI (the regulator), and TnT (the link to the muscle thin filament) have been determined. The structure of troponin in the Ca(2+)-activated state features a nearly twofold symmetrical assembly of TnI and TnT subunits penetrated asymmetrically by the dumbbell-shaped TnC subunit. Ca ions are thought to regulate contraction by controlling the presentation to and withdrawal of the TnI inhibitory segment from the thin filament. Here, we show that the rigid central helix of the sensor binds the inhibitory segment of TnI in the Ca(2+)-activated state. Comparison of crystal structures of troponin in the Ca(2+)-activated state at 3.0 angstroms resolution and in the Ca(2+)-free state at 7.0 angstroms resolution shows that the long framework helices of TnI and TnT, presumed to be a Ca(2+)-independent structural domain of troponin are unchanged. Loss of Ca ions causes the rigid central helix of the sensor to collapse and to release the inhibitory segment of TnI. The inhibitory segment of TnI changes conformation from an extended loop in the presence of Ca2+ to a short alpha-helix in its absence. We also show that Anapoe, a detergent molecule, increases the contractile force of muscle fibers and binds specifically, together with the TnI switch helix, in a hydrophobic pocket of TnC upon activation by Ca ions.
Organizational Affiliation:
Department of Biochemistry and Biophysics, University of California, San Francisco, CA 94143-2240, USA.