Solution structure and dynamics of LuxU from Vibrio harveyi, a phosphotransferase protein involved in bacterial quorum sensing.
Ulrich, D.L., Kojetin, D., Bassler, B.L., Cavanagh, J., Loria, J.P.(2005) J Mol Biol 347: 297-307
- PubMed: 15740742
- DOI: https://doi.org/10.1016/j.jmb.2005.01.039
- Primary Citation of Related Structures:
1Y6D - PubMed Abstract:
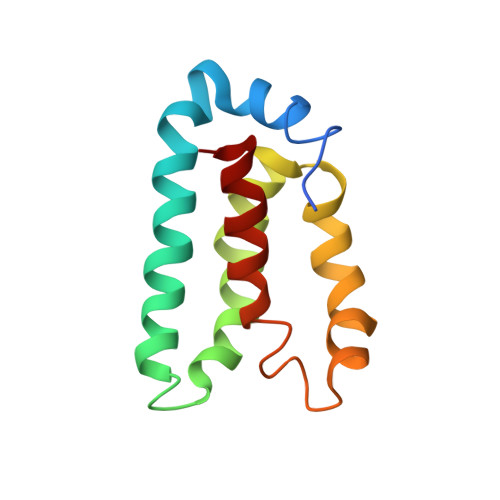
The marine bacterium Vibrio harveyi controls its bioluminescence by a process known as quorum sensing. In this process, autoinducer molecules are detected by membrane-bound sensor kinase/response regulator proteins (LuxN and LuxQ) that relay a signal via a series of protein phosphorylation reactions to another response regulator protein, LuxO. Phosphorylated LuxO indirectly represses the expression of the proteins responsible for bioluminescence. Integral to this quorum sensing process is the function of the phosphotransferase protein, LuxU. LuxU acts to shuttle the phosphate from the membrane-bound proteins, LuxN and LuxQ, to LuxO. LuxU is a 114 amino acid residue monomeric protein. Solution NMR was used to determine the three-dimensional structure of LuxU. LuxU contains a four-helix bundle topology with the active-site histidine residue (His58) located on alpha-helix C and exposed to solution. The active site represents a cluster of positively charged residues located on an otherwise hydrophobic protein face. NMR spin-relaxation experiments identify a collection of flexible residues localized on the same region of LuxU as His58. The studies described here represent the first structural characterization of an isolated, monomeric bacterial phosphotransferase protein.
Organizational Affiliation:
Department of Chemistry, Yale University, P.O. Box 208107, New Haven, CT 06520, USA.