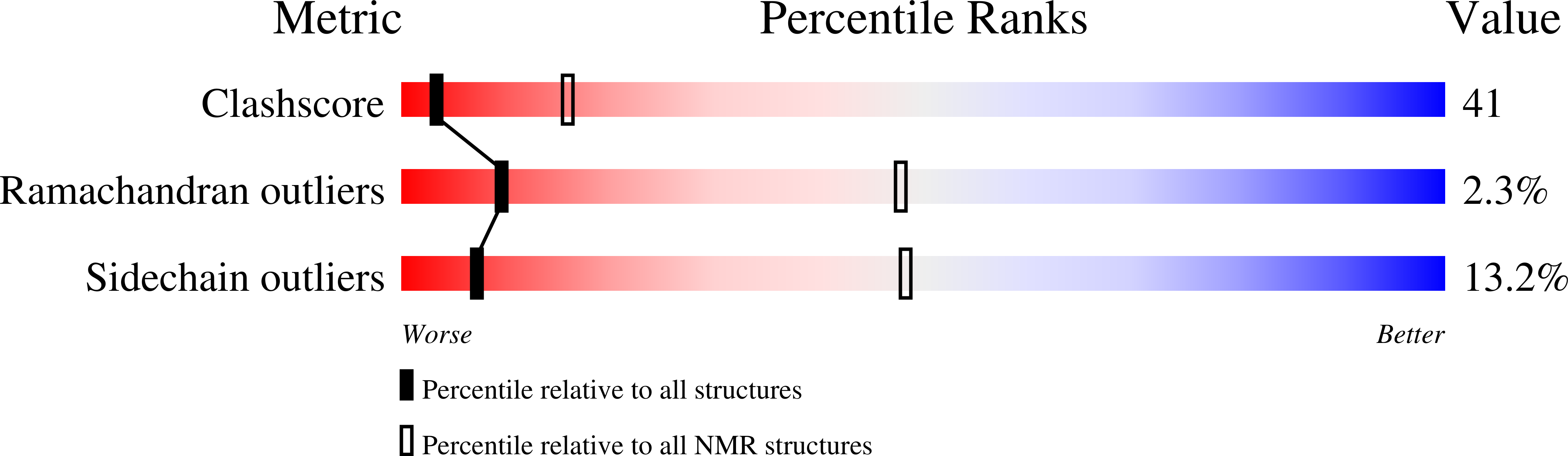The Focal Adhesion Targeting Domain of Focal Adhesion Kinase Contains a Hinge Region that Modulates Tyrosine 926 Phosphorylation.
Prutzman, K.C., Gao, G., King, M.L., Iyer, V.V., Mueller, G.A., Schaller, M.D., Campbell, S.L.(2004) Structure 12: 881-891
- PubMed: 15130480
- DOI: https://doi.org/10.1016/j.str.2004.02.028
- Primary Citation of Related Structures:
1PV3 - PubMed Abstract:
The focal adhesion targeting (FAT) domain of focal adhesion kinase (FAK) is critical for recruitment of FAK to focal adhesions and contains tyrosine 926, which, when phosphorylated, binds the SH2 domain of Grb2. Structural studies have shown that the FAT domain is a four-helix bundle that exists as a monomer and a dimer due to domain swapping of helix 1. Here, we report the NMR solution structure of the avian FAT domain, which is similar in overall structure to the X-ray crystal structures of monomeric forms of the FAT domain, except that loop 1 is longer and less structured in solution. Residues in this region undergo temperature-dependent exchange broadening and sample aberrant phi and psi angles, which suggests that this region samples multiple conformations. We have also identified a mutant that dimerizes approximately 8 fold more than WT FAT domain and exhibits increased phosphorylation of tyrosine 926 both in vitro and in vivo.
Organizational Affiliation:
Department of Biochemistry and Biophysics, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599 USA.