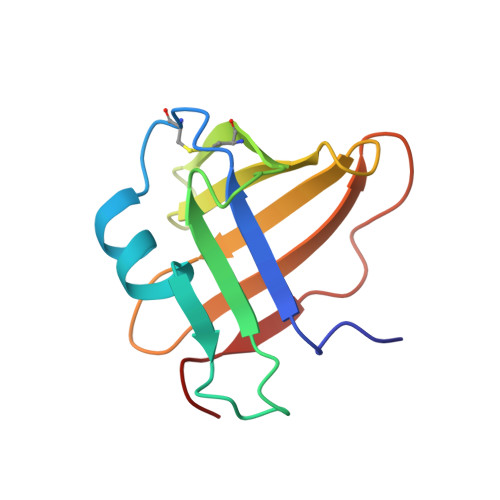
A Novel Class of Cysteine Protease Inhibitors: Solution Structure of Staphostatin a from Staphylococcus Aureus
Dubin, G., Krajewski, M., Popowicz, G., Stec-Niemczyk, J., Bochtler, M., Potempa, J., Dubin, A., Holak, T.A.(2003) Biochemistry 42: 13449
- PubMed: 14621990
- DOI: https://doi.org/10.1021/bi035310j
- Primary Citation of Related Structures:
1OH1 - PubMed Abstract:
A series of secreted proteases are included among the virulence factors documented for Staphylococcus aureus. In light of increasing antibiotic resistance of this dangerous human pathogen, these proteases are considered as suitable targets for the development of novel therapeutic strategies. The recent discovery of staphostatins, endogenous, highly specific, staphylococcal cysteine protease inhibitors, opened a possibility for structure-based design of low molecular weight analogues. Moreover, the crystal structure of staphostatin B revealed a distinct folding pattern and an unexpected, substrate-like binding mode. The solution structure of staphostatin A reported here confirms that staphostatins constitute a novel, distinct class of cysteine protease inhibitors. In addition, the structure knowledge-based mutagenesis studies shed light on individual structural features of staphostatin A, the inhibition mechanism, and the determinants of distinct specificity of staphostatins toward their target proteases.
Organizational Affiliation:
Faculty of Biotechnology, Jagiellonian University, ul. Gronostajowa 7, 30-387 Krakow, Poland. gdubin@mol.uj.edu.pl