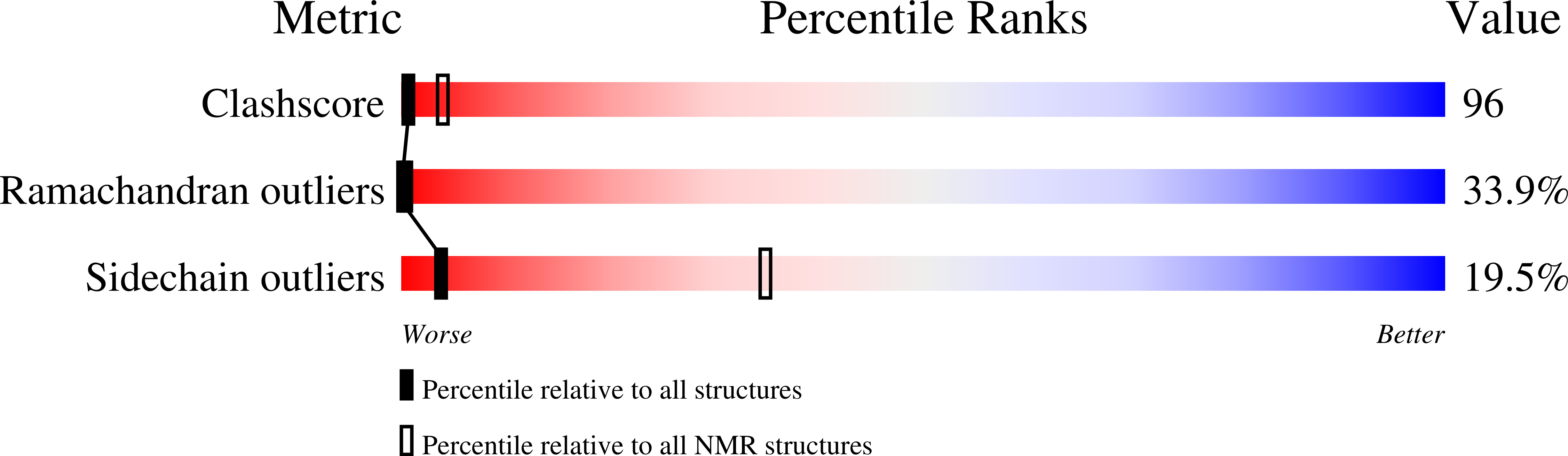Determination of the three-dimensional structure of CC chemokine monocyte chemoattractant protein 3 by 1H two-dimensional NMR spectroscopy.
Meunier, S., Bernassau, J.M., Guillemot, J.C., Ferrara, P., Darbon, H.(1997) Biochemistry 36: 4412-4422
- PubMed: 9109648
- DOI: https://doi.org/10.1021/bi9627929
- Primary Citation of Related Structures:
1NCV - PubMed Abstract:
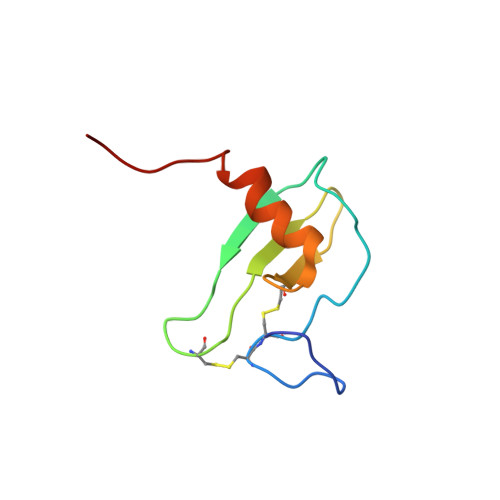
MCP-3 is a beta chemokine consisting of 76 amino acid residues. It has been described to be involved in the activation of all leukocytic cells, activation mediated by the presence of multiple binding sites on the target cells. Its three-dimensional structure has been studied by making use of two-dimensional 1H NMR spectroscopy. MCP-3 exhibits the same monomeric structure as the other chemokines, i.e., a three-stranded antiparallel beta sheet covered on one face by an alpha helix. Although it belongs to the same subfamily as RANTES (Chung et al., 1995; Faitbrother et al., 1994) and hMIP-1beta (Lodi et al., 1994), the MCP-3 dimer is folded like IL-8 with the so-called alphabeta sandwich structural motif. Structural and sequence analysis gives clear indications suggesting that the other MCP chemokines may have the same quaternary structure, contrary to the other beta chemokines.
Organizational Affiliation:
AFMB-IFR1, UPR 9039-CNRS, Marseille, France.