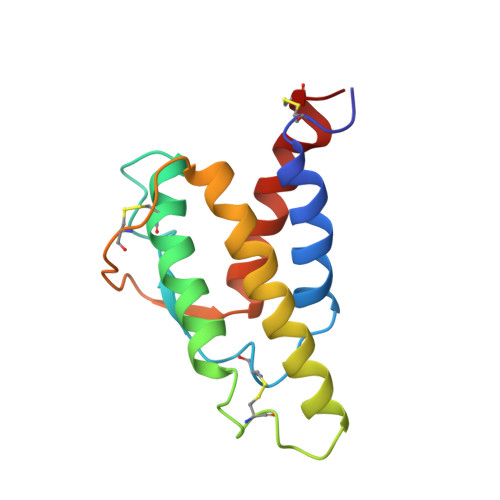
Human interleukin 4. The solution structure of a four-helix bundle protein.
Smith, L.J., Redfield, C., Boyd, J., Lawrence, G.M., Edwards, R.G., Smith, R.A., Dobson, C.M.(1992) J Mol Biol 224: 899-904
- PubMed: 1569578
- DOI: https://doi.org/10.1016/0022-2836(92)90457-u
- Primary Citation of Related Structures:
1ITL - PubMed Abstract:
Heteronuclear 13C and 15N three-dimensional nuclear magnetic resonance (n.m.r.) techniques have been used to determine the solution structure of human interleukin 4, a four-helix bundle protein. A dynamical simulated annealing protocol was used to calculate an ensemble of structures from an n.m.r. data set of 1735 distance restraints, 101 phi angle restraints and 27 pairs of hydrogen bond restraints. The protein structure has a left-handed up-up-down-down topology for the four helices with the two long overhand loops in the structure being connected by a short section of irregular antiparallel beta-sheet. Analysis of the side-chains in the protein shows a clustering of hydrophobic residues, particularly leucines, in the core of the bundle with the side-chains of charged residues being located on the protein surface. The solution structure has been compared with a recent structure prediction for human interleukin 4 and with crystal structures of other helix bundle proteins.
Organizational Affiliation:
Inorganic Chemistry Laboratory, University of Oxford, England.