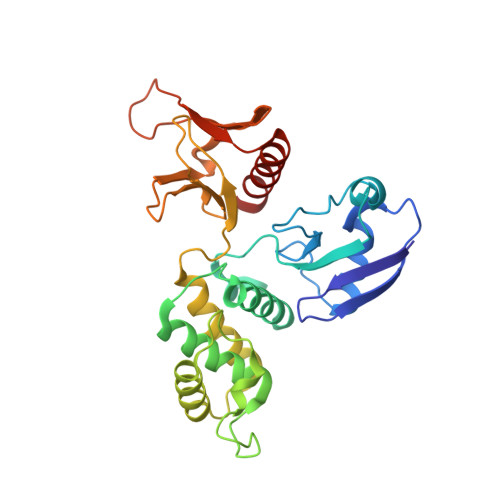
Protein 4.1R core domain structure and insights into regulation of cytoskeletal organization.
Han, B.G., Nunomura, W., Takakuwa, Y., Mohandas, N., Jap, B.K.(2000) Nat Struct Biol 7: 871-875
- PubMed: 11017195
- DOI: https://doi.org/10.1038/82819
- Primary Citation of Related Structures:
1GG3 - PubMed Abstract:
The crystal structure of the core domain (N-terminal 30 kDa domain) of cytoskeletal protein 4.1R has been determined and shows a cloverleaf-like architecture. Each lobe of the cloverleaf contains a specific binding site for either band 3, glycophorin C/D or p55. At a central region of the molecule near where the three lobes are joined are two separate calmodulin (CaM) binding regions. One of these is composed primarily of an alpha-helix and is Ca 2+ insensitive; the other takes the form of an extended structure and its binding with CaM is dramatically enhanced by the presence of Ca 2+, resulting in the weakening of protein 4.1R binding to its target proteins. This novel architecture, in which the three lobes bind with three membrane associated proteins, and the location of calmodulin binding sites provide insight into how the protein 4.1R core domain interacts with membrane proteins and dynamically regulates cell shape in response to changes in intracellular Ca2+ levels.
Organizational Affiliation:
Life Sciences Division, Lawrence Berkeley National Laboratory, University of California, Berkeley, California 94720, USA.