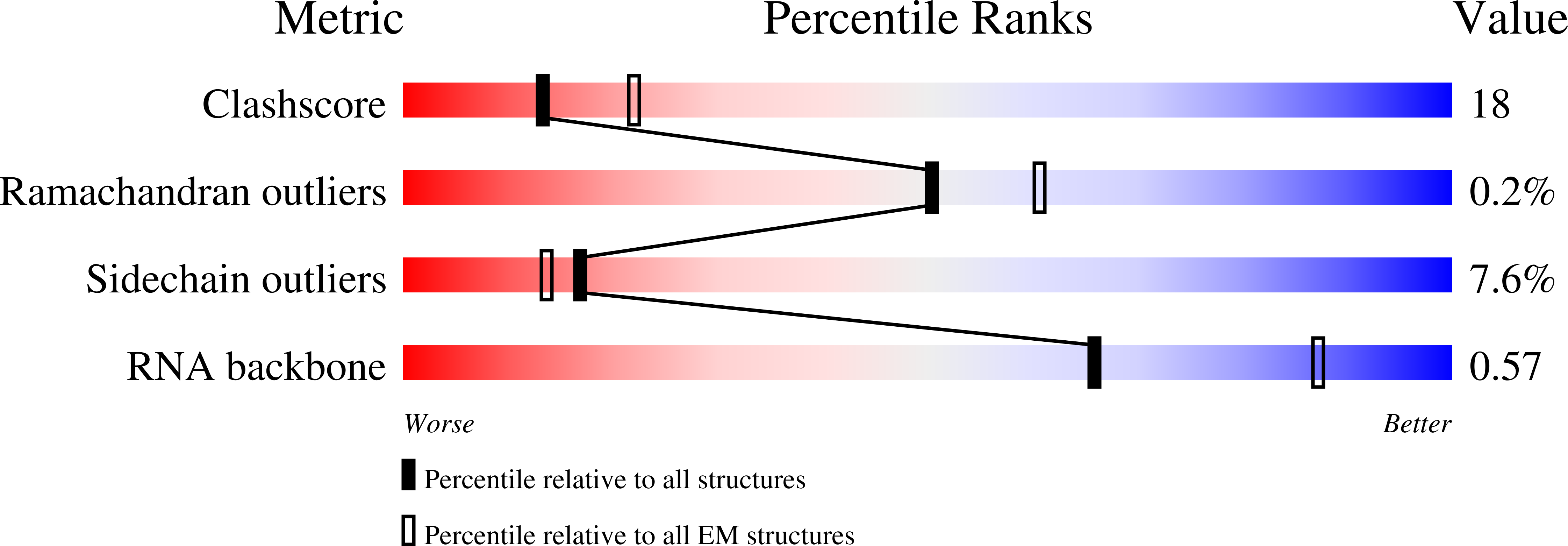Structural basis for HIV-1 antagonism of host APOBEC3G via Cullin E3 ligase.
Ito, F., Alvarez-Cabrera, A.L., Liu, S., Yang, H., Shiriaeva, A., Zhou, Z.H., Chen, X.S.(2023) Sci Adv 9: eade3168-eade3168
- PubMed: 36598981
- DOI: https://doi.org/10.1126/sciadv.ade3168
- Primary Citation of Related Structures:
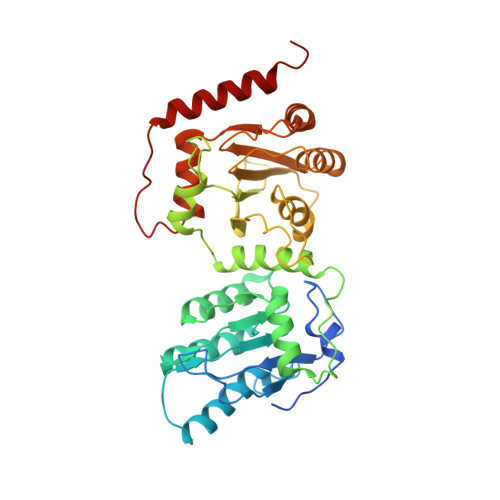
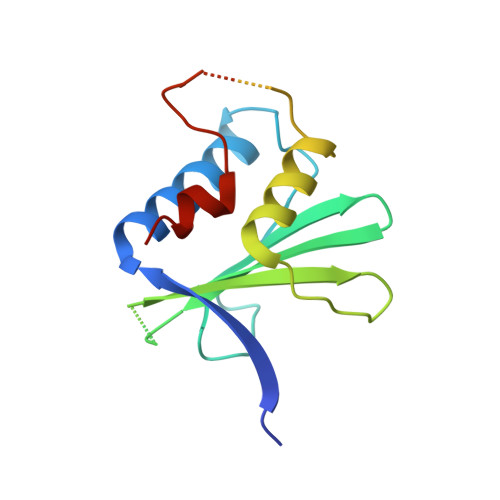
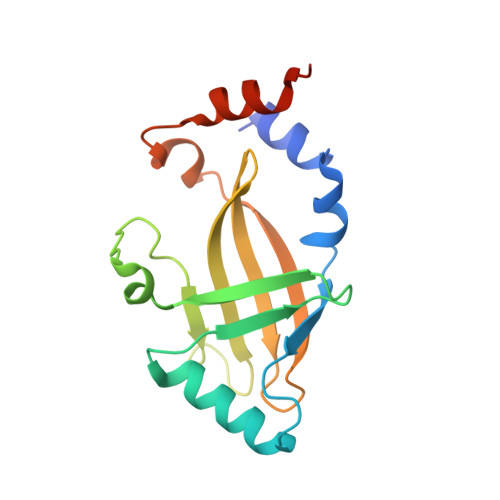
8E40 - PubMed Abstract:
Human APOBEC3G (A3G) is a virus restriction factor that inhibits HIV-1 replication and triggers lethal hypermutation on viral reverse transcripts. HIV-1 viral infectivity factor (Vif) breaches this host A3G immunity by hijacking a cellular E3 ubiquitin ligase complex to target A3G for ubiquitination and degradation. The molecular mechanism of A3G targeting by Vif-E3 ligase is unknown, limiting the antiviral efforts targeting this host-pathogen interaction crucial for HIV-1 infection. Here, we report the cryo-electron microscopy structures of A3G bound to HIV-1 Vif in complex with T cell transcription cofactor CBF-β and multiple components of the Cullin-5 RING E3 ubiquitin ligase. The structures reveal unexpected RNA-mediated interactions of Vif with A3G primarily through A3G's noncatalytic domain, while A3G's catalytic domain is poised for ubiquitin transfer. These structures elucidate the molecular mechanism by which HIV-1 Vif hijacks the host ubiquitin ligase to specifically target A3G to establish infection and offer structural information for the rational development of antiretroviral therapeutics.
Organizational Affiliation:
Molecular and Computational Biology, Departments of Biological Sciences, University of Southern California, Los Angeles, CA, USA.