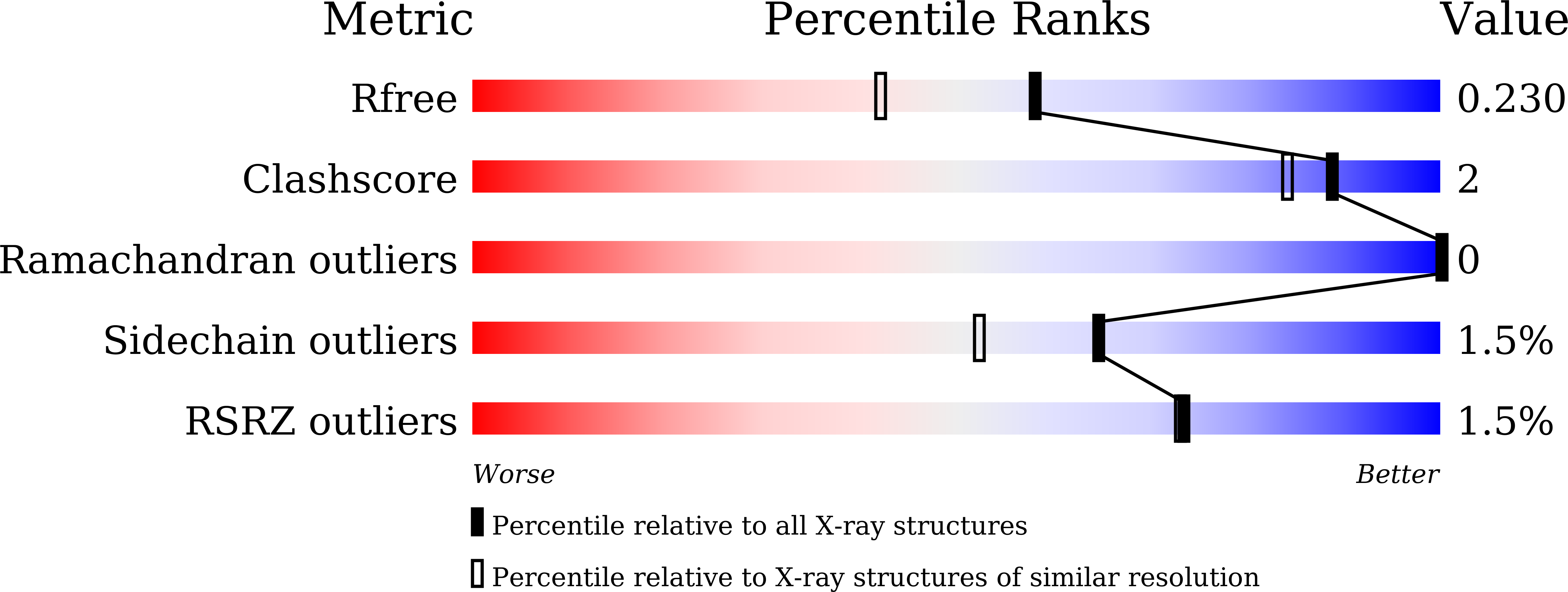
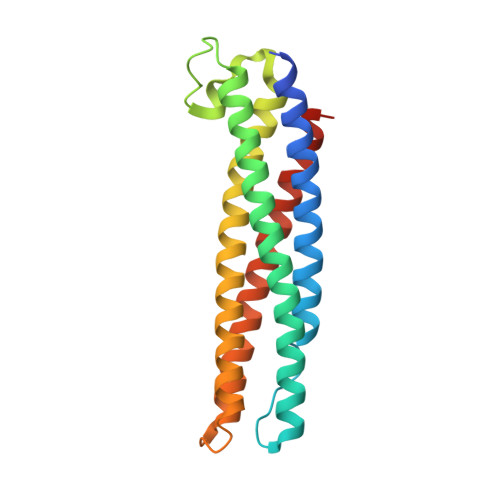
Cooperative regulation of PBI1 and MAPKs controls WRKY45 transcription factor in rice immunity.
Ichimaru, K., Yamaguchi, K., Harada, K., Nishio, Y., Hori, M., Ishikawa, K., Inoue, H., Shigeta, S., Inoue, K., Shimada, K., Yoshimura, S., Takeda, T., Yamashita, E., Fujiwara, T., Nakagawa, A., Kojima, C., Kawasaki, T.(2022) Nat Commun 13: 2397-2397
- PubMed: 35577789
- DOI: https://doi.org/10.1038/s41467-022-30131-y
- Primary Citation of Related Structures:
7X8V - PubMed Abstract:
The U-box type ubiquitin ligase PUB44 positively regulates pattern-triggered immunity in rice. Here, we identify PBI1, a protein that interacts with PUB44. Crystal structure analysis indicates that PBI1 forms a four-helix bundle structure. PBI1 also interacts with WRKY45, a master transcriptional activator of rice immunity, and negatively regulates its activity. PBI1 is degraded upon perception of chitin, and this is suppressed by silencing of PUB44 or expression of XopP, indicating that PBI1 degradation depends on PUB44. These data suggest that PBI1 suppresses WRKY45 activity when cells are in an unelicited state, and during chitin signaling, PUB44-mediated degradation of PBI1 leads to activation of WRKY45. In addition, chitin-induced MAP kinase activation is required for WRKY45 activation and PBI1 degradation. These results demonstrate that chitin-induced activation of WRKY45 is regulated by the cooperation between MAP kinase-mediated phosphorylation and PUB44-mediated PBI1 degradation.
Organizational Affiliation:
Department of Advanced Bioscience, Graduate School of Agriculture, Kindai University, Nakamachi, Nara, 631-8505, Japan.