Holliday junction resolution by At-HIGLE: an SLX1 lineage endonuclease from Arabidopsis thaliana with a novel in-built regulatory mechanism.
Verma, P., Kumari, P., Negi, S., Yadav, G., Gaur, V.(2022) Nucleic Acids Res 50: 4630-4646
- PubMed: 35412622
- DOI: https://doi.org/10.1093/nar/gkac239
- Primary Citation of Related Structures:
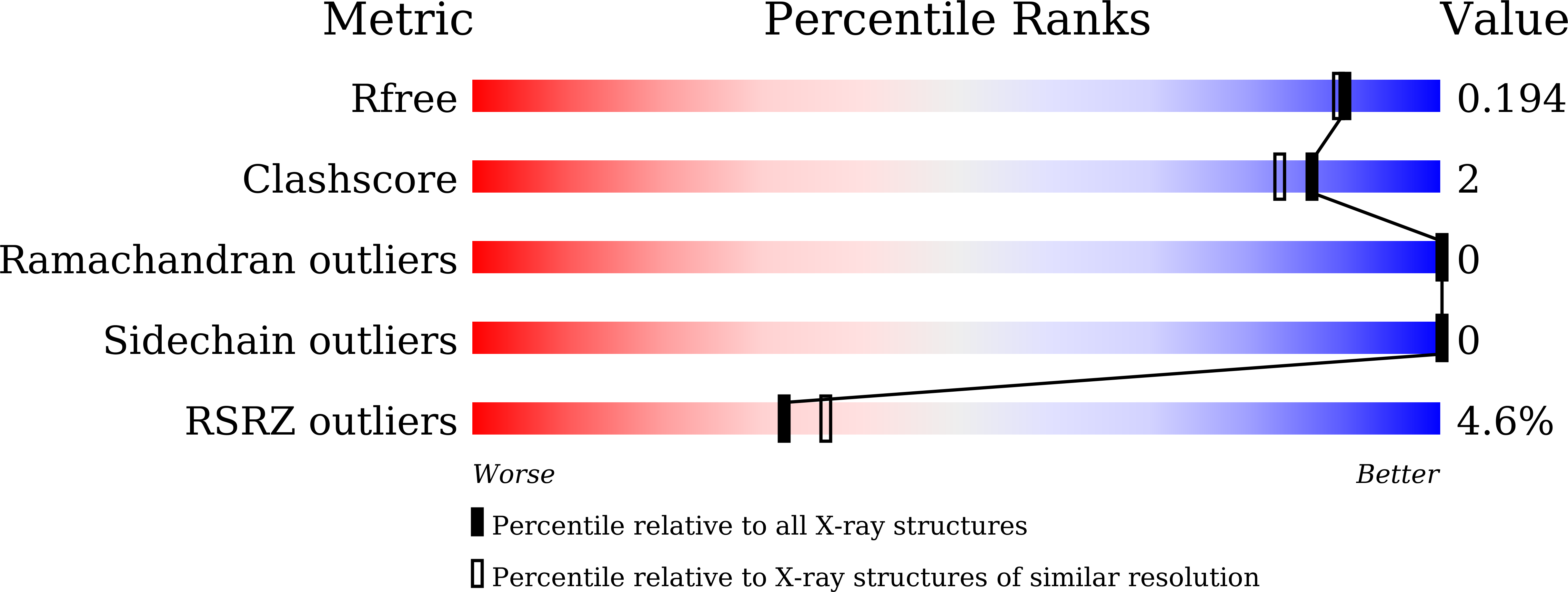
7WME - PubMed Abstract:
Holliday junction is the key homologous recombination intermediate, resolved by structure-selective endonucleases (SSEs). SLX1 is the most promiscuous SSE of the GIY-YIG nuclease superfamily. In fungi and animals, SLX1 nuclease activity relies on a non-enzymatic partner, SLX4, but no SLX1-SLX4 like complex has ever been characterized in plants. Plants exhibit specialized DNA repair and recombination machinery. Based on sequence similarity with the GIY-YIG nuclease domain of SLX1 proteins from fungi and animals, At-HIGLE was identified to be a possible SLX1 like nuclease from plants. Here, we elucidated the crystal structure of the At-HIGLE nuclease domain from Arabidopsis thaliana, establishing it as a member of the SLX1-lineage of the GIY-YIG superfamily with structural changes in DNA interacting regions. We show that At-HIGLE can process branched-DNA molecules without an SLX4 like protein. Unlike fungal SLX1, At-HIGLE exists as a catalytically active homodimer capable of generating two coordinated nicks during HJ resolution. Truncating the extended C-terminal region of At-HIGLE increases its catalytic activity, changes the nicking pattern, and monomerizes At-HIGLE. Overall, we elucidated the first structure of a plant SLX1-lineage protein, showed its HJ resolving activity independent of any regulatory protein, and identified an in-built novel regulatory mechanism engaging its C-terminal region.
Organizational Affiliation:
National Institute of Plant Genome Research, Aruna Asaf Ali Marg, New Delhi 110067, India.