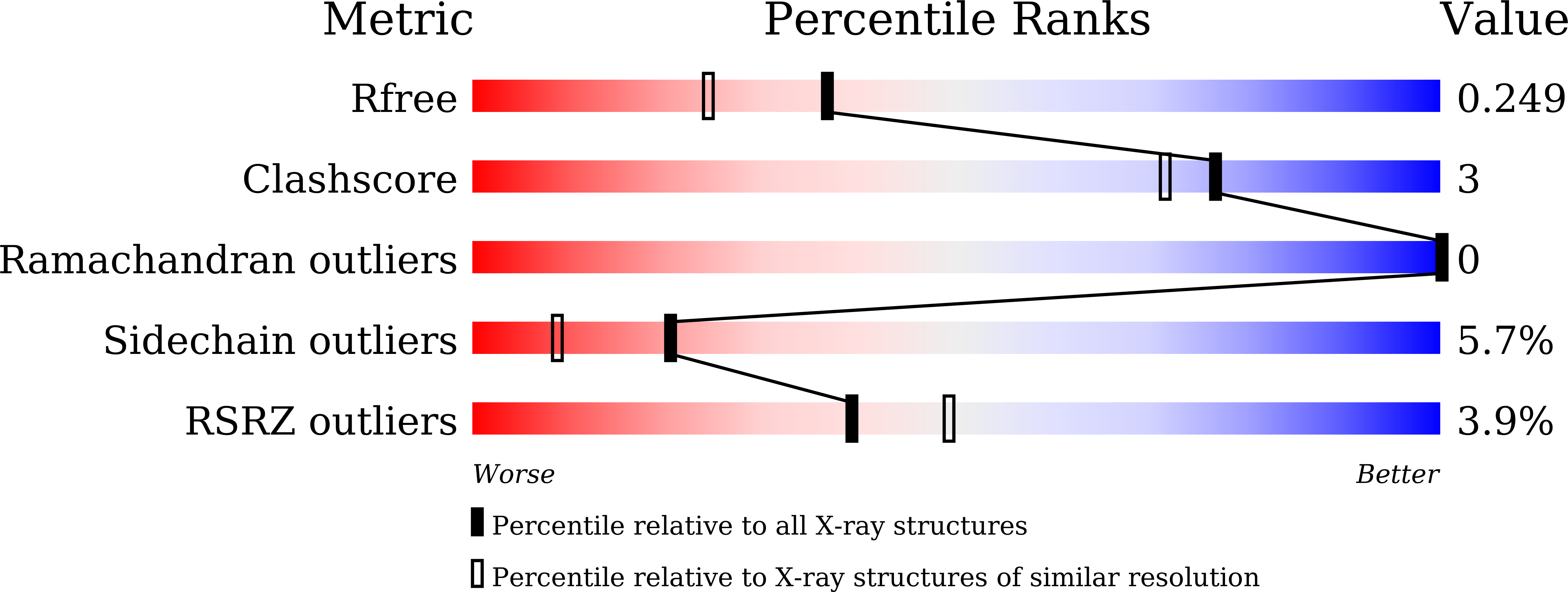
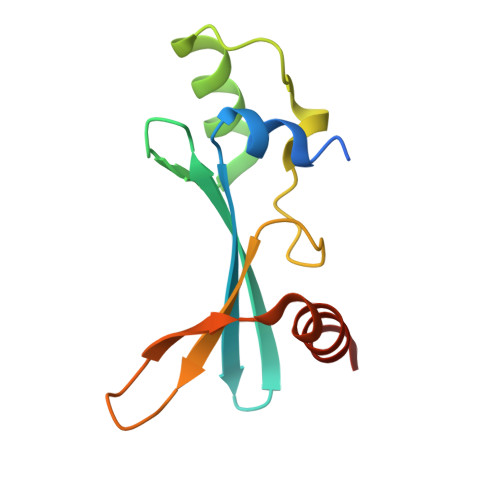
Crystal structure of the BAZ2B TAM domain.
Feng, Y., Chen, S., Zhou, M., Zhang, J., Min, J., Liu, K.(2022) Heliyon 8: e09873-e09873
- PubMed: 35865993
- DOI: https://doi.org/10.1016/j.heliyon.2022.e09873
- Primary Citation of Related Structures:
7MWH, 7WIN - PubMed Abstract:
BAZ2B is a regulatory subunit of the ISWI (Imitation Switch) remodeling complex and engages in nucleosome remodeling. Loss-of-function and haploinsufficiency of BAZ2B are associated with different diseases. BAZ2B is a large multidomain protein. In addition to the epigenetic reader domains plant homeodomain (PHD) and bromodomain (BRD), BAZ2B also has a Tip5/ARBP/MBD (TAM) domain. Sequence alignment revealed that the TAM domains of BAZ2A and BAZ2B share 53% sequence identity. How the BAZ2A TAM domain bound with DNA has been characterized recently, however, the DNA binding ability and methylation preference, as well as the structural basis of the BAZ2B TAM domain are not studied yet. In this study, we measured the DNA binding affinity of the TAM domain of BAZ2B, and also determined its apo crystal structure. We found that the TAM domains of BAZ2A and BAZ2B adopt almost the same fold, and like BAZ2A, the BAZ2B TAM domain also binds to dsDNA without methyl-cytosine preference, implying that the BAZ2B TAM domain might recognize DNA in a similar binding mode to that of the BAZ2A TAM domain. These results provide clues for the biological function study of BAZ2B in the future.
Organizational Affiliation:
Hubei Key Laboratory of Genetic Regulation and Integrative Biology, School of Life Sciences, Central China Normal University, Wuhan, 430079, PR China.