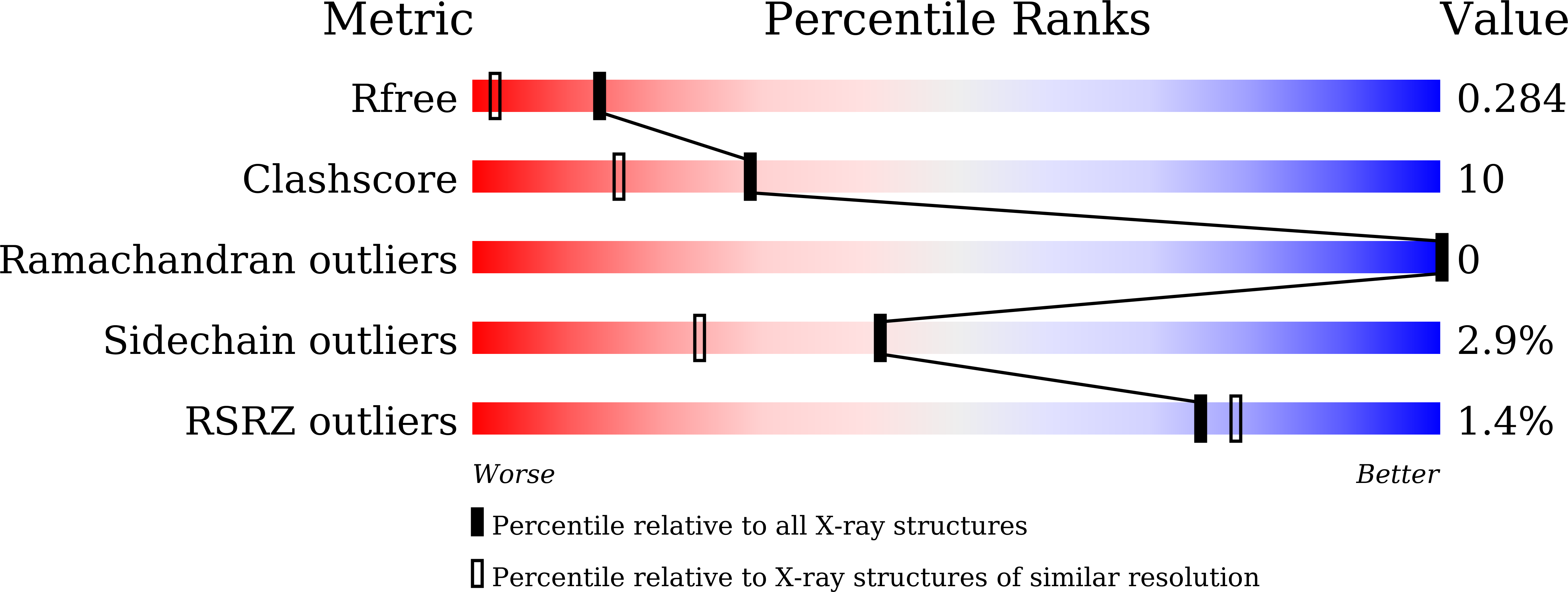
Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Zhang, K., Li, Z., Zhu, Q., Cao, H., He, X., Zhang, X.H., Liu, W., Lyu, Q.(2022) Commun Biol 5: 782-782
- PubMed: 35918517
- DOI: https://doi.org/10.1038/s42003-022-03721-1
- Primary Citation of Related Structures:
7W12, 7W13, 7W16, 7W18 - PubMed Abstract:
Alginate lyases can be used to produce well-defined alginate oligosaccharides (AOSs) because of their specificities for AOS products. A large number of alginate lyases have been recorded in the CAZy database; however, the majority are annotated-only alginate lyases that include little information on their products, thus limiting their applications. Here, we establish a simple and experiment-saving approach to predict product distributions for PL7 alginate lyases through extensive structural biology, bioinformatics and biochemical studies. Structural study on several PL7 alginate lyases reveals that two loops around the substrate binding cleft determine product distribution. Furthermore, a database containing the loop information of all annotated-only single-domain PL7 alginate lyases is constructed, enabling systematic exploration of the association between loop and product distribution. Based on these results, a simplified loop/product distribution relationship is proposed, giving us information on product distribution directly from the amino acid sequence.
Organizational Affiliation:
MOE Key Laboratory of Marine Genetics and Breeding, College of Marine Life Sciences, Ocean University of China, Qingdao, China.