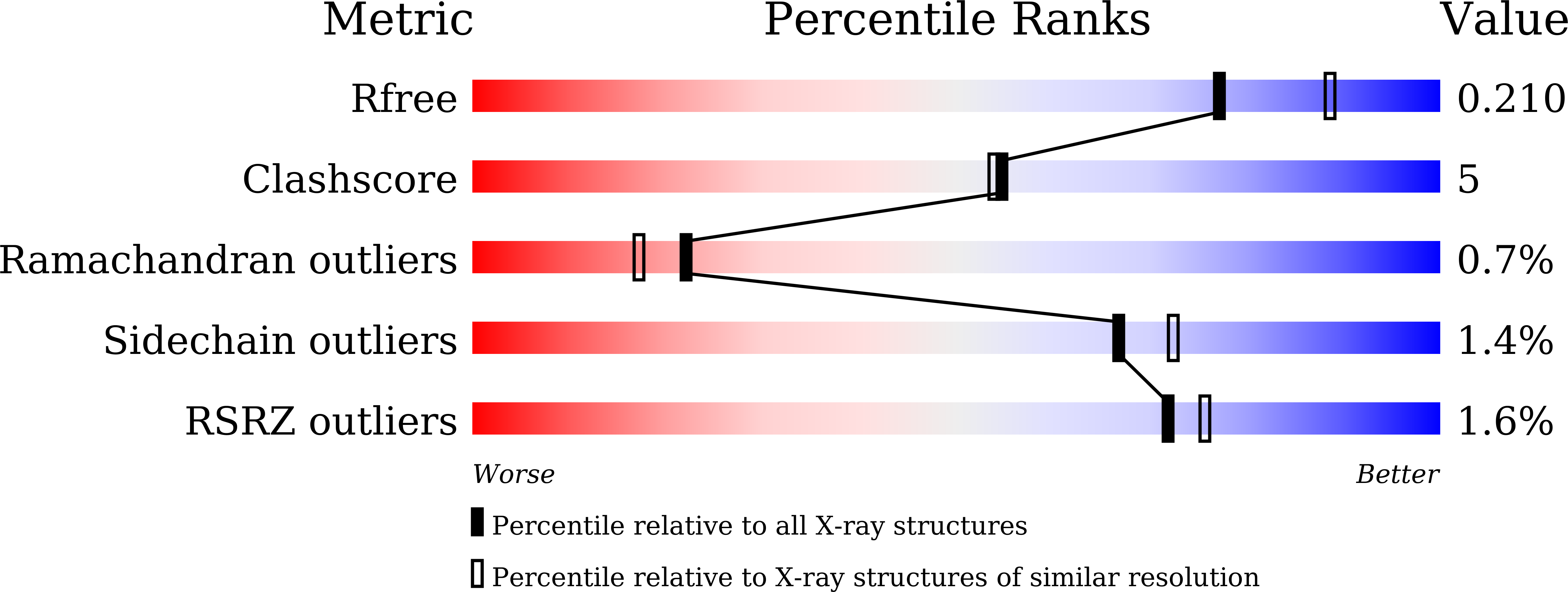
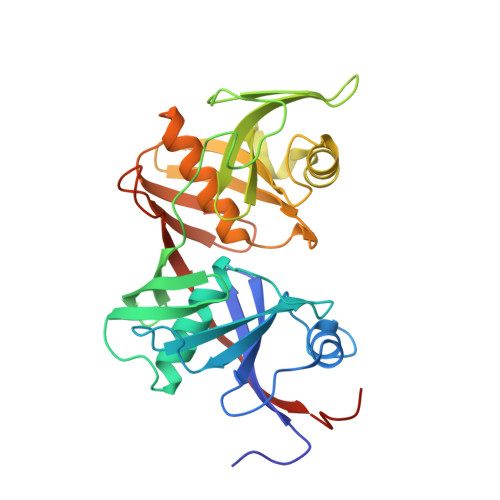
Crystal structure of O-ureidoserine racemase found in the d-cycloserine biosynthetic pathway.
Oda, K., Sakaguchi, T., Matoba, Y.(2022) Proteins 90: 912-918
- PubMed: 34877716
- DOI: https://doi.org/10.1002/prot.26290
- Primary Citation of Related Structures:
7VDY - PubMed Abstract:
The O-ureidoserine racemase (DcsC) is an enzyme found from the biosynthetic gene cluster of antitubercular agent d-cycloserine. Although DcsC is homologous to diaminopimelate epimerase (DapF) that catalyzes the interconversion between ll- and dl-diaminopimelic acid, it specifically catalyzes the interconversion between O-ureido-l-serine and its enantiomer. Here we determined the crystal structure of DcsC at a resolution of 2.12 Å, implicating that the catalytic mechanism of DcsC shares similarity with that of DapF. Comparing the structure of the active center of DcsC to that of DapF, Thr72, Thr198, and Tyr219 of DcsC are likely to be involved in the substrate specificity.
Organizational Affiliation:
Department of Virology, Institute of Biomedical and Health Sciences, Hiroshima University, Hiroshima, Japan.