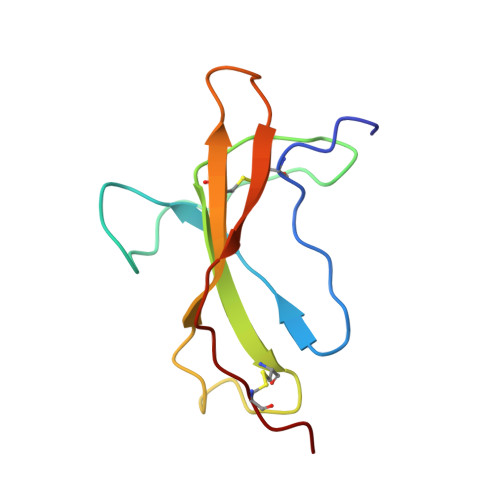
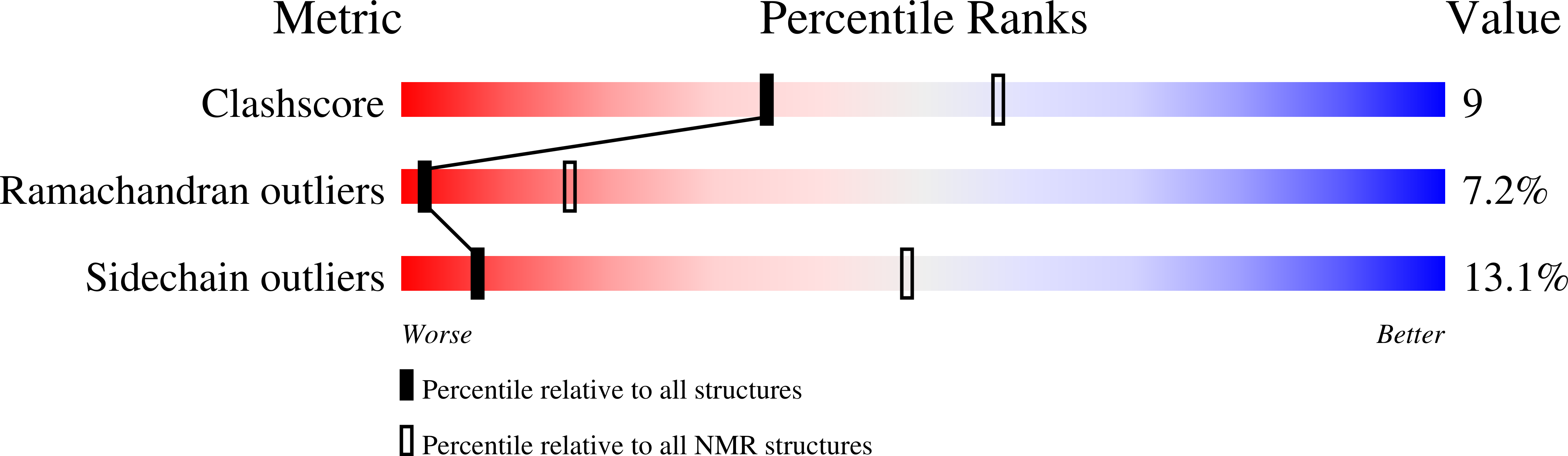Convergent evolution of a parasite-encoded complement control protein-scaffold to mimic binding of mammalian TGF-beta to its receptors, T beta RI and T beta RII.
Mukundan, A., Byeon, C.H., Hinck, C.S., Cunningham, K., Campion, T., Smyth, D.J., Maizels, R.M., Hinck, A.P.(2022) J Biol Chem 298: 101994-101994
- PubMed: 35500648
- DOI: https://doi.org/10.1016/j.jbc.2022.101994
- Primary Citation of Related Structures:
7SXB - PubMed Abstract:
The mouse intestinal helminth Heligmosomoides polygyrus modulates host immune responses by secreting a transforming growth factor (TGF)-β mimic (TGM), to expand the population of Foxp3 + T regs . TGM comprises five complement control protein (CCP)-like domains, designated D1-D5. Though lacking homology to TGF-β, TGM binds directly to the TGF-β receptors TβRI and TβRII and stimulates the differentiation of naïve T-cells into T regs . However, the molecular determinants of binding are unclear. Here, we used surface plasmon resonance, isothermal calorimetry, NMR spectroscopy, and mutagenesis to investigate how TGM binds the TGF-β receptors. We demonstrate that binding is modular, with D1-D2 binding to TβRI and D3 binding to TβRII. D1-D2 and D3 were further shown to compete with TGF-β(TβRII) 2 and TGF-β for binding to TβRI and TβRII, respectively. The solution structure of TGM-D3 revealed that TGM adopts a CCP-like fold but is also modified to allow the C-terminal strand to diverge, leading to an expansion of the domain and opening potential interaction surfaces. TGM-D3 also incorporates a long structurally ordered hypervariable loop, adding further potential interaction sites. Through NMR shift perturbations and binding studies of TGM-D3 and TβRII variants, TGM-D3 was shown to occupy the same site of TβRII as bound by TGF-β using both a novel interaction surface and the hypervariable loop. These results, together with the identification of other secreted CCP-like proteins with immunomodulatory activity in H. polygyrus, suggest that TGM is part of a larger family of evolutionarily plastic parasite effector molecules that mediate novel interactions with their host.
Organizational Affiliation:
Department of Structural Biology, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania USA.