Borrelia miyamotoi FbpA and FbpB Are Immunomodulatory Outer Surface Lipoproteins With Distinct Structures and Functions.
Booth Jr., C.E., Powell-Pierce, A.D., Skare, J.T., Garcia, B.L.(2022) Front Immunol 13: 886733-886733
- PubMed: 35693799
- DOI: https://doi.org/10.3389/fimmu.2022.886733
- Primary Citation of Related Structures:
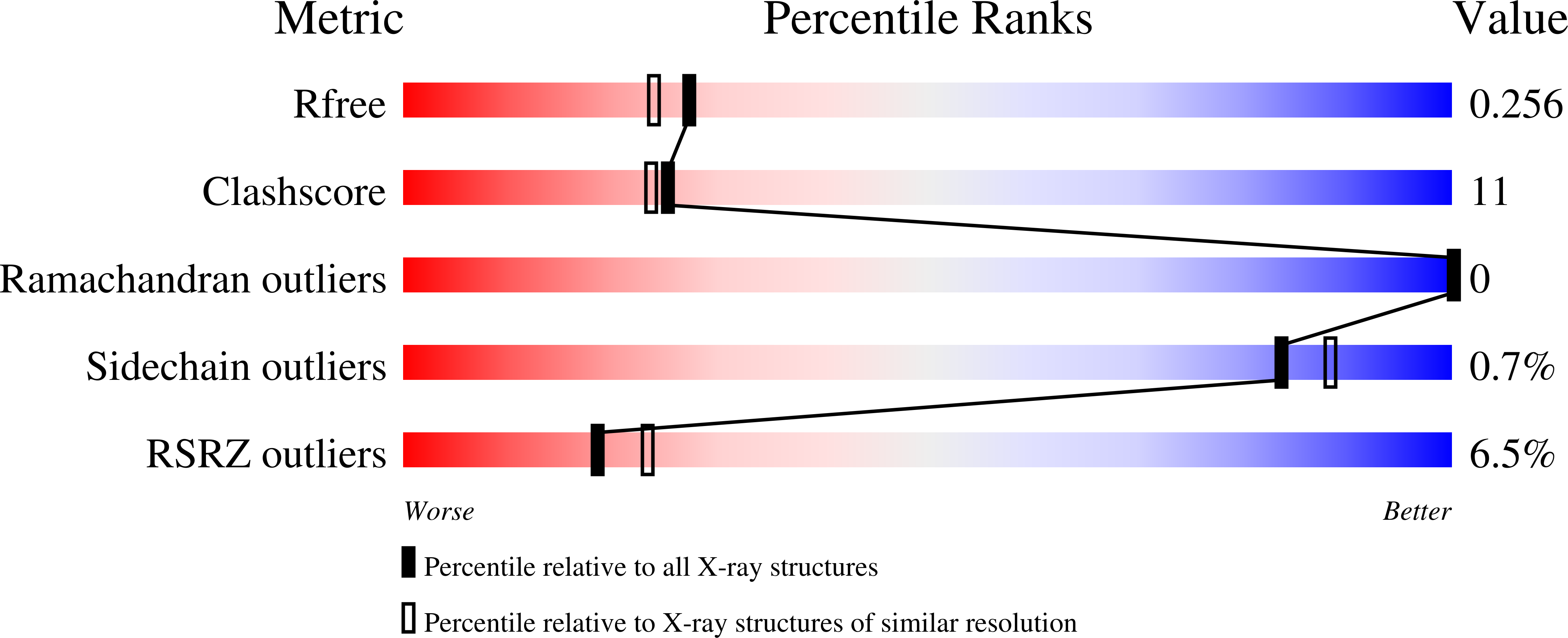
7RPR, 7RPS - PubMed Abstract:
Pathogens that traffic in the blood of their hosts must employ mechanisms to evade the host innate immune system, including the complement cascade. The Lyme disease spirochete, Borreliella burgdorferi , has evolved numerous outer membrane lipoproteins that interact directly with host proteins. Compared to Lyme disease-associated spirochetes, relatively little is known about how an emerging tick-borne spirochetal pathogen, Borrelia miyamotoi , utilizes surface lipoproteins to interact with a human host. B. burgdorferi expresses the multifunctional lipoprotein, BBK32, that inhibits the classical pathway of complement through interaction with the initiating protease C1r, and also interacts with fibronectin using a separate intrinsically disordered domain. B. miyamotoi encodes two separate bbk32 orthologs denoted fbpA and fbpB ; however, the activities of these proteins are unknown. Here, we show that B. miyamotoi FbpA binds human fibronectin in a manner similar to B. burgdorferi BBK32, whereas FbpB does not. FbpA and FbpB both bind human complement C1r and protect a serum-sensitive B. burgdorferi strain from complement-mediated killing, but surprisingly, differ in their ability to recognize activated C1r versus zymogen states of C1r. To better understand the observed differences in C1r recognition and inhibition properties, high-resolution X-ray crystallography structures were solved of the C1r-binding regions of B. miyamotoi FbpA and FbpB at 1.9Å and 2.1Å, respectively. Collectively, these data suggest that FbpA and FbpB have partially overlapping functions but are functionally and structurally distinct. The data presented herein enhances our overall understanding of how bloodborne pathogens interact with fibronectin and modulate the complement system.
Organizational Affiliation:
Department of Microbiology and Immunology, Brody School of Medicine, East Carolina University, Greenville, NC, United States.