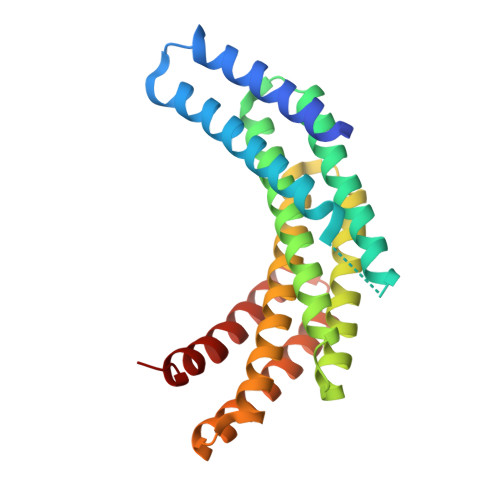
De novo design of protein homodimers containing tunable symmetric protein pockets.
Hicks, D.R., Kennedy, M.A., Thompson, K.A., DeWitt, M., Coventry, B., Kang, A., Bera, A.K., Brunette, T.J., Sankaran, B., Stoddard, B., Baker, D.(2022) Proc Natl Acad Sci U S A 119: e2113400119-e2113400119
- PubMed: 35862457
- DOI: https://doi.org/10.1073/pnas.2113400119
- Primary Citation of Related Structures:
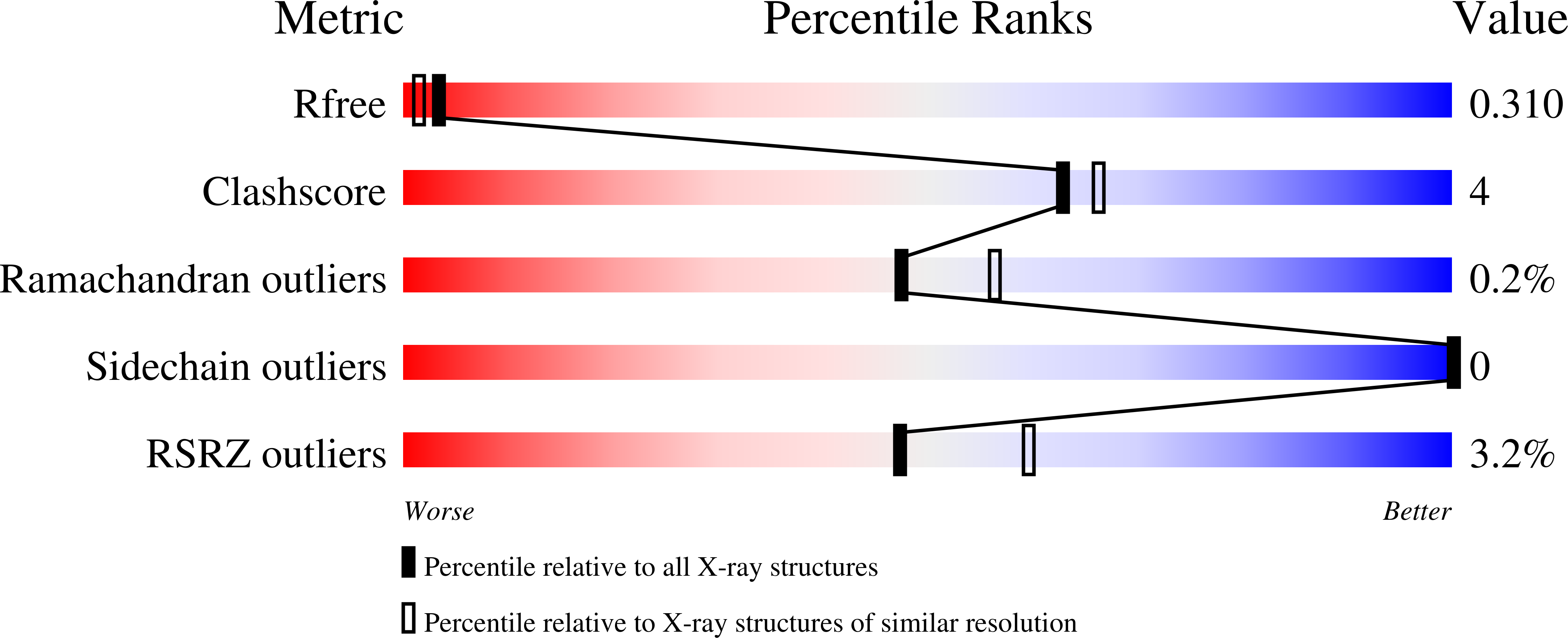
7RKC, 7RMX, 7RMY - PubMed Abstract:
Function follows form in biology, and the binding of small molecules requires proteins with pockets that match the shape of the ligand. For design of binding to symmetric ligands, protein homo-oligomers with matching symmetry are advantageous as each protein subunit can make identical interactions with the ligand. Here, we describe a general approach to designing hyperstable C2 symmetric proteins with pockets of diverse size and shape. We first designed repeat proteins that sample a continuum of curvatures but have low helical rise, then docked these into C2 symmetric homodimers to generate an extensive range of C2 symmetric cavities. We used this approach to design thousands of C2 symmetric homodimers, and characterized 101 of them experimentally. Of these, the geometry of 31 were confirmed by small angle X-ray scattering and 2 were shown by crystallographic analyses to be in close agreement with the computational design models. These scaffolds provide a rich set of starting points for binding a wide range of C2 symmetric compounds.
Organizational Affiliation:
Department of Biochemistry, University of Washington, Seattle, WA 98195.