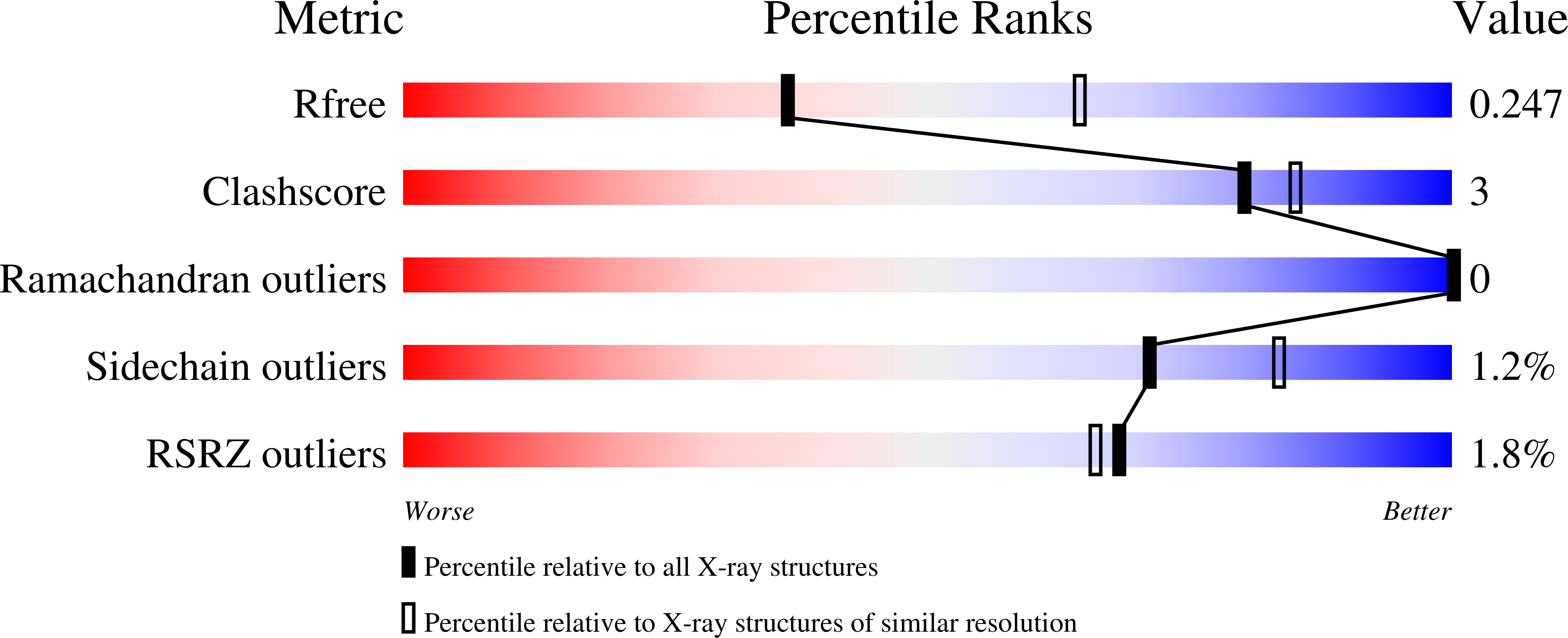
The suppressor of copper sensitivity protein C from Caulobacter crescentus is a trimeric disulfide isomerase that binds copper(I) with subpicomolar affinity.
Petit, G.A., Hong, Y., Djoko, K.Y., Whitten, A.E., Furlong, E.J., McCoy, A.J., Gulbis, J.M., Totsika, M., Martin, J.L., Halili, M.A.(2022) Acta Crystallogr D Struct Biol 78: 337-352
- PubMed: 35234148
- DOI: https://doi.org/10.1107/S2059798322000729
- Primary Citation of Related Structures:
7RGV - PubMed Abstract:
The introduction of disulfide bonds into periplasmic proteins is a critical process in many Gram-negative bacteria. The formation and regulation of protein disulfide bonds have been linked to the production of virulence factors. Understanding the different pathways involved in this process is important in the development of strategies to disarm pathogenic bacteria. The well characterized disulfide bond-forming (DSB) proteins play a key role by introducing or isomerizing disulfide bonds between cysteines in substrate proteins. Curiously, the suppressor of copper sensitivity C proteins (ScsCs), which are part of the bacterial copper-resistance response, share structural and functional similarities with DSB oxidase and isomerase proteins, including the presence of a catalytic thioredoxin domain. However, the oxidoreductase activity of ScsC varies with its oligomerization state, which depends on a poorly conserved N-terminal domain. Here, the structure and function of Caulobacter crescentus ScsC (CcScsC) have been characterized. It is shown that CcScsC binds copper in the copper(I) form with subpicomolar affinity and that its isomerase activity is comparable to that of Escherichia coli DsbC, the prototypical dimeric bacterial isomerase. It is also reported that CcScsC functionally complements trimeric Proteus mirabilis ScsC (PmScsC) in vivo, enabling the swarming of P. mirabilis in the presence of copper. Using mass photometry and small-angle X-ray scattering (SAXS) the protein is demonstrated to be trimeric in solution, like PmScsC, and not dimeric like EcDsbC. The crystal structure of CcScsC was also determined at a resolution of 2.6 Å, confirming the trimeric state and indicating that the trimerization results from interactions between the N-terminal α-helical domains of three CcScsC protomers. The SAXS data analysis suggested that the protomers are dynamic, like those of PmScsC, and are able to sample different conformations in solution.
Organizational Affiliation:
Griffith Institute for Drug Discovery, Griffith University, Don Young Road, Nathan, QLD 4111, Australia.