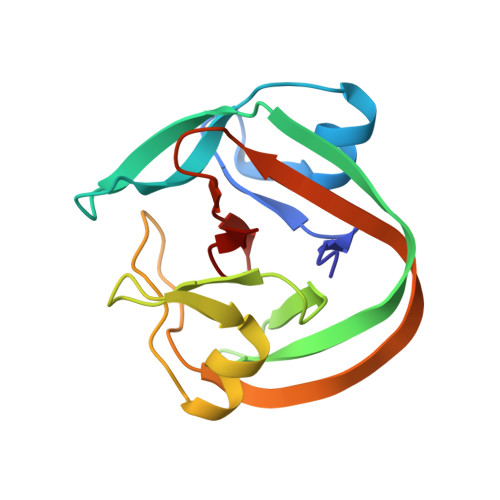
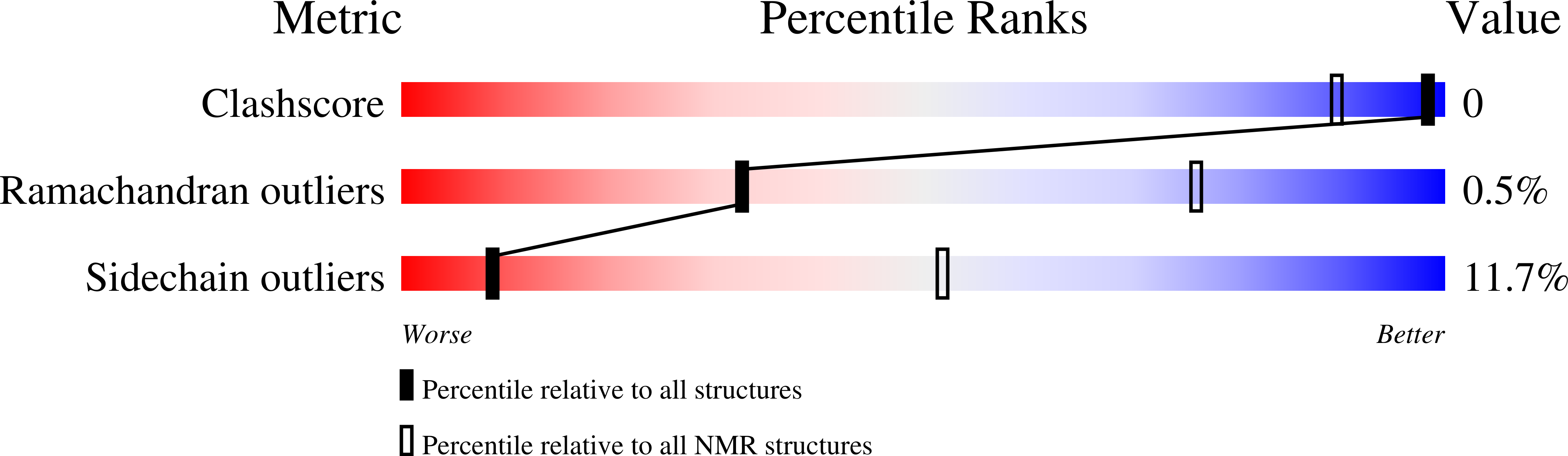The NMR structure of the engineered halophilic DnaE intein for segmental isotopic labeling using conditional protein splicing.
Heikkinen, H.A., Aranko, A.S., Iwai, H.(2022) J Magn Reson (1969 338: 107195-107195
- PubMed: 35398651
- DOI: https://doi.org/10.1016/j.jmr.2022.107195
- Primary Citation of Related Structures:
7QIL - PubMed Abstract:
Protein trans-splicing catalyzed by split inteins has been used for segmental isotopic labeling of proteins for alleviating the complexity of NMR signals. Whereas inteins spontaneously trigger protein splicing upon protein folding, inteins from extremely halophilic organisms require a high salinity condition to induce protein splicing. We designed and created a salt-inducible intein from the widely used DnaE intein from Nostoc punctiforme by introducing 29 mutations, which required a lower salt concentration than naturally occurring halo-obligate inteins. We determined the NMR solution structure of the engineered salt-inducible DnaE intein in 2 M NaCl, showing the essentially identical three-dimensional structure to the original one, albeit it unfolds without salts. The NMR structure of a halo-obligate intein under high salinity suggests that the stabilization of the active folded conformation is not a mere result of various intramolecular interactions but the subtle energy balance from the complex interactions, including the solvation energy, which involve waters, ions, co-solutes, and protein polypeptide chains.
Organizational Affiliation:
Institute of Biotechnology, University of Helsinki, PO Box 65, Helsinki, FIN-00014, Finland.