Comparative structural analysis provides new insights into the function of R2-like ligand-binding oxidase.
Diamanti, R., Srinivas, V., Johansson, A.I., Nordstrom, A., Griese, J.J., Lebrette, H., Hogbom, M.(2022) FEBS Lett 596: 1600-1610
- PubMed: 35175627
- DOI: https://doi.org/10.1002/1873-3468.14319
- Primary Citation of Related Structures:
7QBK, 7QBP - PubMed Abstract:
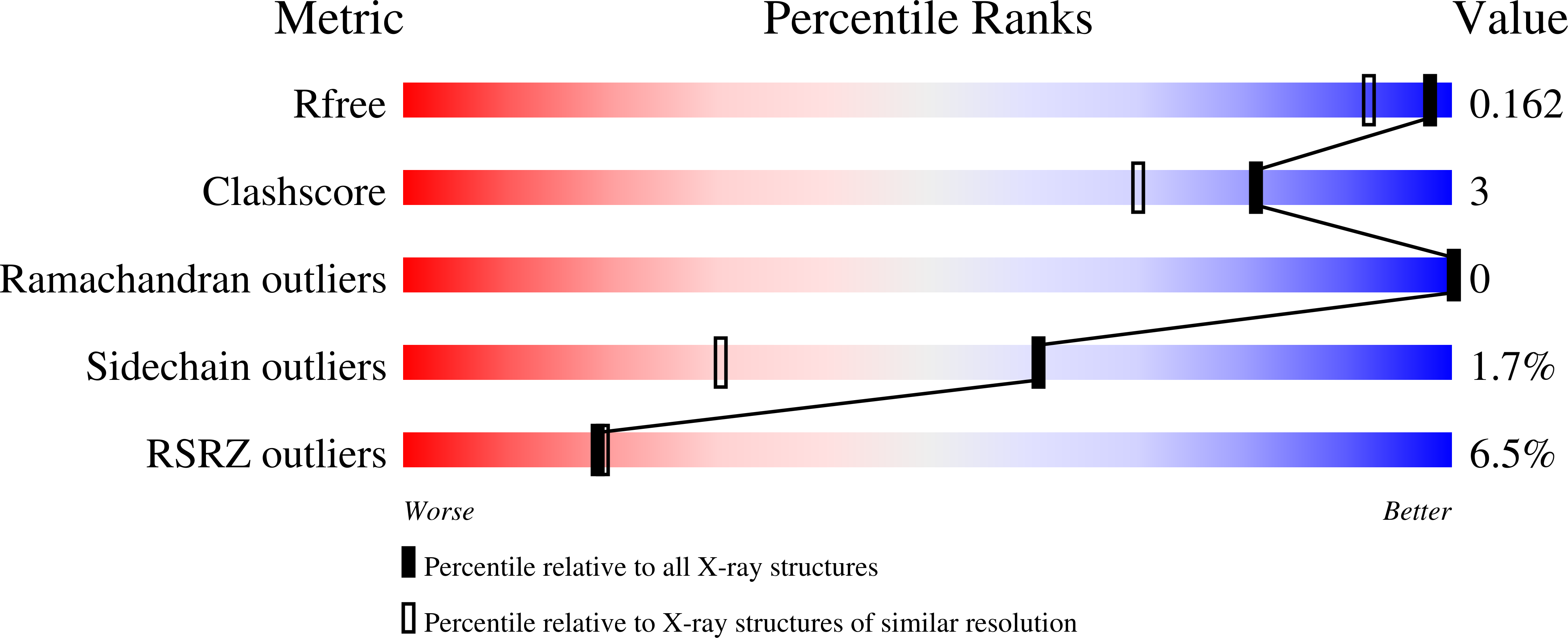
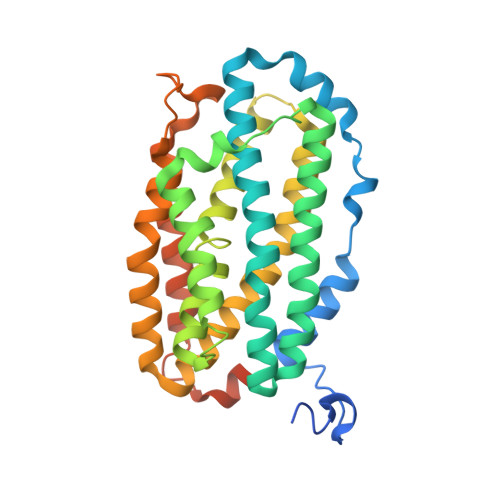
R2-like ligand-binding oxidase (R2lox) is a ferritin-like protein that harbours a heterodinuclear manganese-iron active site. Although R2lox function is yet to be established, the enzyme binds a fatty acid ligand coordinating the metal centre and catalyses the formation of a tyrosine-valine ether cross-link in the protein scaffold upon O 2 activation. Here, we characterized the ligands copurified with R2lox by mass spectrometry-based metabolomics. Moreover, we present the crystal structures of two new homologs of R2lox, from Saccharopolyspora erythraea and Sulfolobus acidocaldarius, at 1.38 Å and 2.26 Å resolution, respectively, providing the highest resolution structure for R2lox, as well as new insights into putative mechanisms regulating the function of the enzyme.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Stockholm University, Stockholm, Sweden.