Deciphering the structure of Arabidopsis thaliana 5- enol -pyruvyl-shikimate-3-phosphate synthase: An essential step toward the discovery of novel inhibitors to supersede glyphosate.
Ruszkowski, M., Forlani, G.(2022) Comput Struct Biotechnol J 20: 1494-1505
- PubMed: 35422967
- DOI: https://doi.org/10.1016/j.csbj.2022.03.020
- Primary Citation of Related Structures:
7PXY - PubMed Abstract:
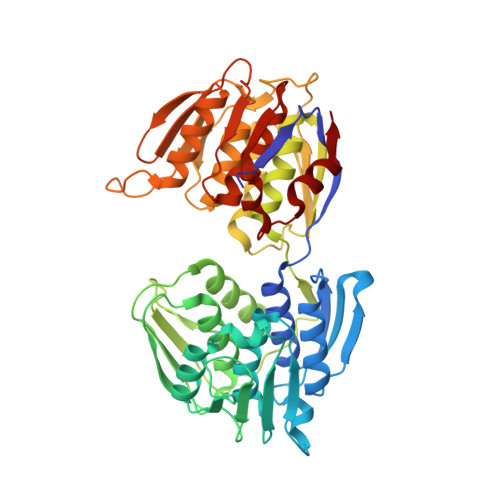
Glyphosate interferes with plant aromatic metabolism through the inhibition of 5- enol -pyruvyl-shikimate-3-phosphate (EPSP) synthase [EPSPS, EC 2.5.1.19]. For this reason, EPSPS has been extensively studied in a vast array of organisms. This notwithstanding, up to date, the crystal structure of the protein has been solved exclusively in a few prokaryotes, while that of the plant enzyme has been only deduced in silico by similarity. This study aimed at determining the structure of EPSPS from the plant model species Arabidopsis thaliana , which has been cloned, heterologously expressed and affinity-purified. The kinetic properties of the enzyme have been determined, as well as its susceptibility to the inhibition brought about by glyphosate. The crystal structure of the protein has been resolved at high resolution (1.4 Å), showing open conformation of the enzyme, which is the state ready for substrate/inhibitor binding. This provides a framework for the structure-based design of novel EPSPS inhibitors. Surface regions near the active-site cleft entrance or at the interdomain hinge appear promising for inhibitor selectivity, while bound chloride near the active site is a potential placeholder for anionic moieties of future herbicides.
Organizational Affiliation:
Department of Structural Biology of Eukaryotes, Institute of Bioorganic Chemistry, Polish Academy of Sciences, Poznan, Poland.