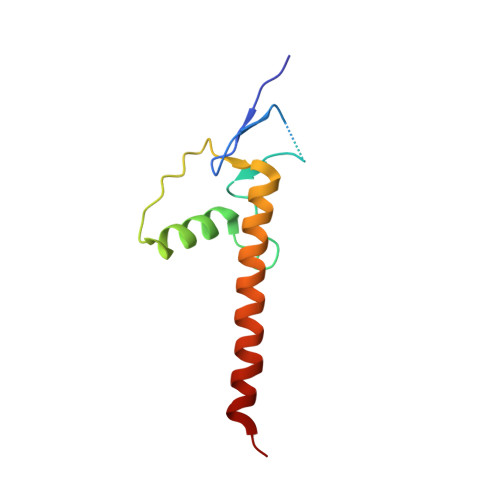
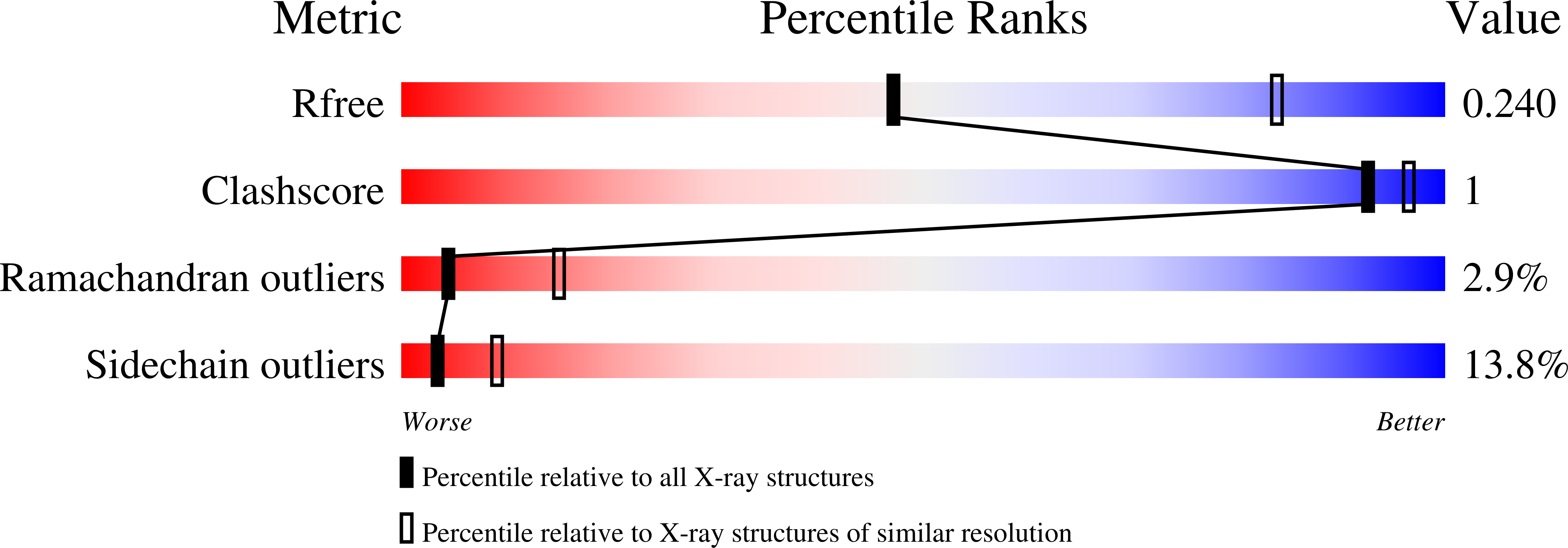Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Bonchuk, A.N., Boyko, K.M., Nikolaeva, A.Y., Burtseva, A.D., Popov, V.O., Georgiev, P.G.(2022) Structure 30: 1004
- PubMed: 35580610
- DOI: https://doi.org/10.1016/j.str.2022.04.009
- Primary Citation of Related Structures:
7PO9, 7POH, 7POK, 7PPP - PubMed Abstract:
ZAD is a C4 zinc-coordinating domain often found at the N-terminus mostly of arthropodan transcription factors with multiple C2H2 zinc-finger domains involved in the regulation of chromosome architecture and promotor activity. ZADs predominantly form homodimers and have low primary sequence similarity. We obtained three crystal structures of the most phylogenetically distant Drosophila ZADs and structure of the only known ZAD-like domain from a mammalian protein (ZNF276). All ZAD structures demonstrate unity of the spatial fold as well as some unique structural features. The specific homodimerization of ZAD is primarily determined by the position and size of secondary structural elements and is further strengthened by a number of unique interactions between subunits. Structural comparison allowed for unraveling key sequence features underlying the similarity of the spatial fold. These features result in a broad variety of ZADs in Arthropod C2H2 proteins, allowing for the emergence of a wide range of highly specific homodimers.
Organizational Affiliation:
Department of the Control of Genetic Processes, Institute of Gene Biology Russian Academy of Sciences, 34/5 Vavilov St., Moscow 119334, Russia; Center for Precision Genome Editing and Genetic Technologies for Biomedicine, Institute of Gene Biology, Russian Academy of Sciences, 34/5 Vavilov St., Moscow 119334, Russia. Electronic address: bonchuk_a@genebiology.ru.