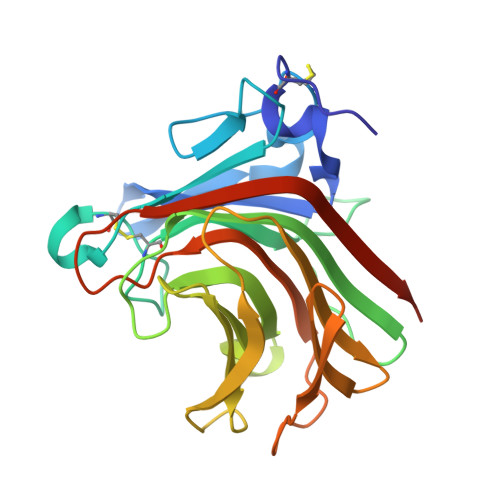
Glucuronan lyases from family PL7 use a Tyr/Tyr syn beta-elimination catalytic mechanism for glucuronan breakdown.
Vuillemin, M., Pilgaard, B., Kiehn, E., Fredslund, F., Welner, D.H., Meyer, A.S., Aachmann, F.L., Wilkens, C.(2024) Chem Commun (Camb) 60: 440-443
- PubMed: 38087900
- DOI: https://doi.org/10.1039/d3cc04256a
- Primary Citation of Related Structures:
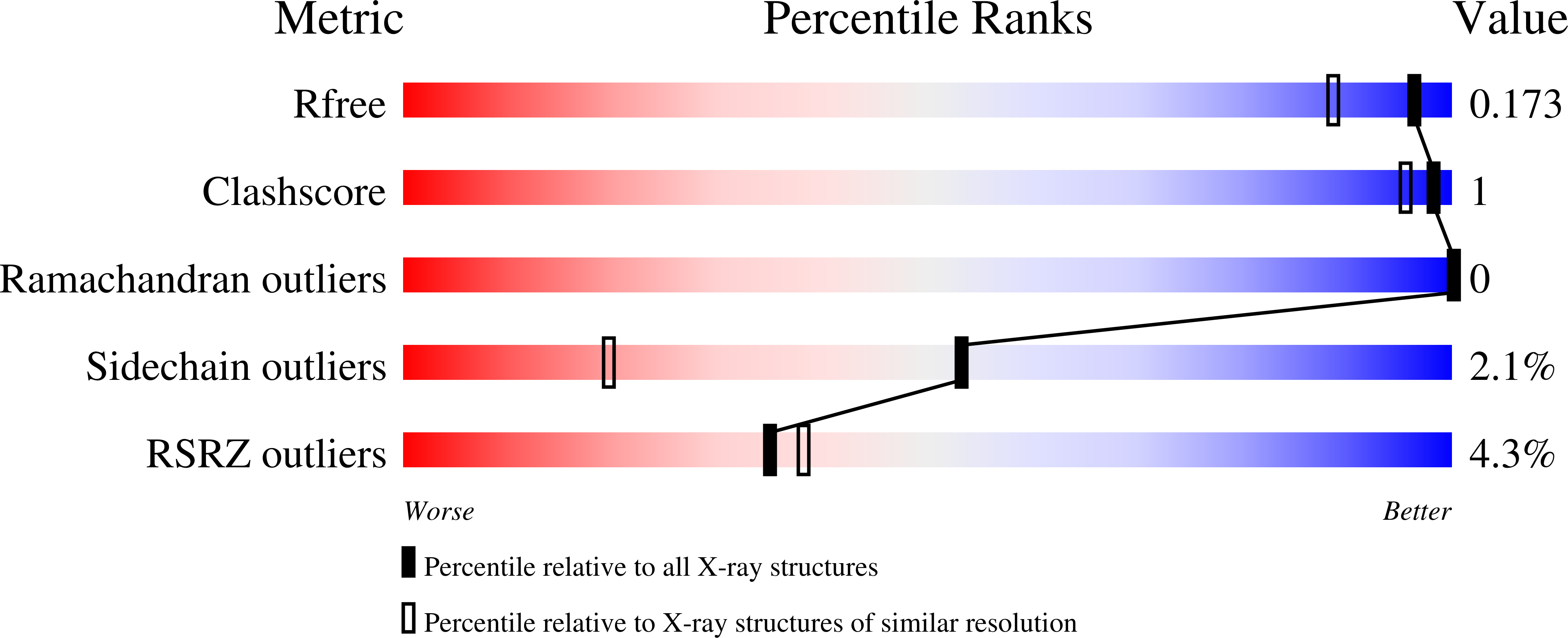
7NDE - PubMed Abstract:
TpPL7A and TpPL7B, members of CAZy family PL7, act as β-glucuronan lyases. TpPL7A diverges by lacking the catalytic histidine, identified as the Brønsted base in PL7 alginate lyases. Our research, including TpPL7A's crystal structure, and mutagenesis studies, reveals a shared syn -β-elimination mechanism with a single tyrosine serving as both base and acid catalyst. This mechanism may extend to subfamily PL7_4 glucuronan lyases.
Organizational Affiliation:
Department of Biotechnology and Biomedicine, Technical University of Denmark, Søltofts Plads, Building 224, DK-2800 Kgs. Lyngby, Denmark. cpwk@novonordisk.com.