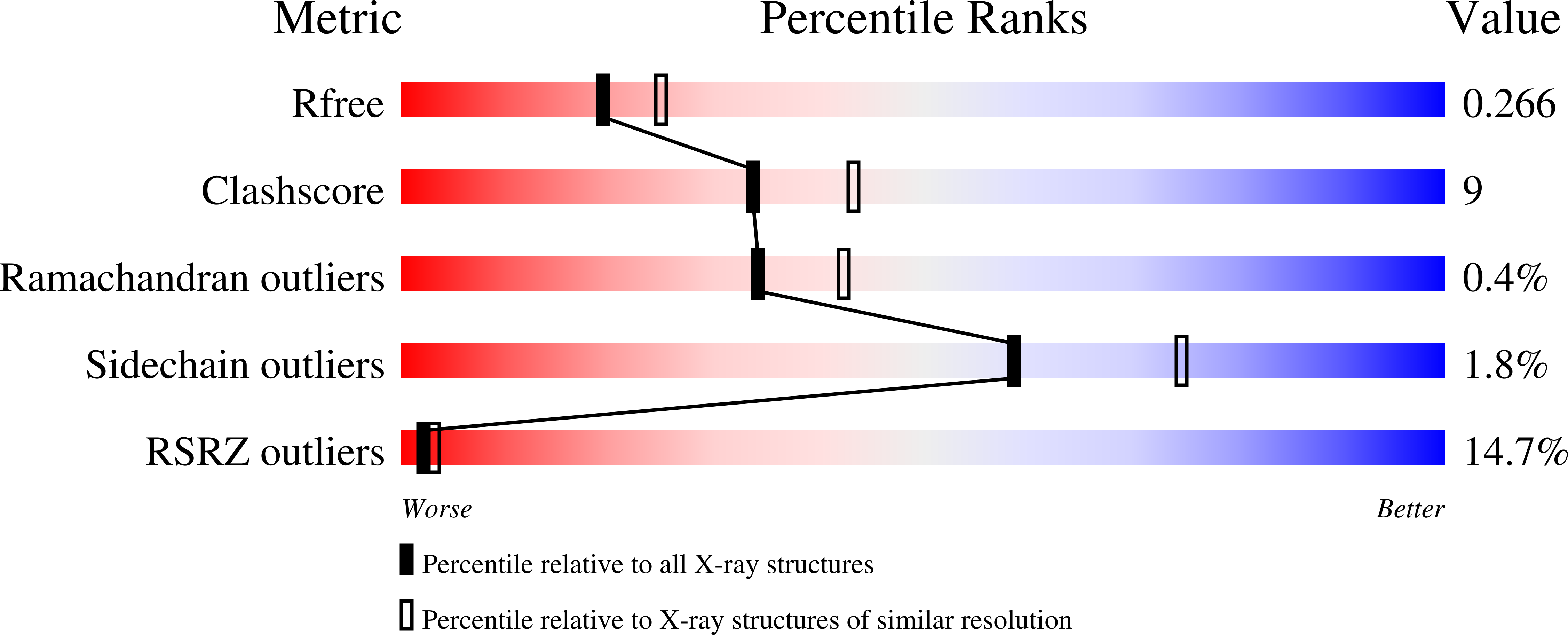
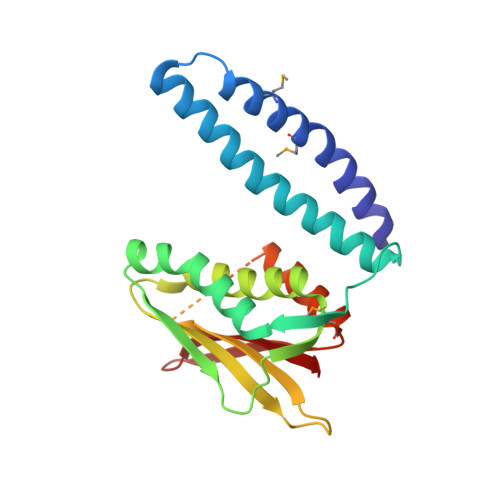
RetS inhibits Pseudomonas aeruginosa biofilm formation by disrupting the canonical histidine kinase dimerization interface of GacS.
Ryan Kaler, K.M., Nix, J.C., Schubot, F.D.(2021) J Biol Chem 297: 101193-101193
- PubMed: 34529974
- DOI: https://doi.org/10.1016/j.jbc.2021.101193
- Primary Citation of Related Structures:
7N0E - PubMed Abstract:
Bacterial signaling histidine kinases (HKs) have long been postulated to function exclusively through linear signal transduction chains. However, several HKs have recently been shown to form complex multikinase networks (MKNs). The most prominent MKN, involving the enzymes RetS and GacS, controls the switch between the motile and biofilm lifestyles in the pathogenic bacterium Pseudomonas aeruginosa. While GacS promotes biofilm formation, RetS counteracts GacS using three distinct mechanisms. Two are dephosphorylating mechanisms. The third, a direct binding between the RetS and GacS HK regions, blocks GacS autophosphorylation. Focusing on the third mechanism, we determined the crystal structure of a cocomplex between the HK region of RetS and the dimerization and histidine phosphotransfer (DHp) domain of GacS. This is the first reported structure of a complex between two distinct bacterial signaling HKs. In the complex, the canonical HK homodimerization interface is replaced by a strikingly similar heterodimeric interface between RetS and GacS. We further demonstrate that GacS autophosphorylates in trans, thus explaining why the formation of a RetS-GacS complex inhibits GacS autophosphorylation. Using mutational analysis in conjunction with bacterial two-hybrid and biofilm assays, we not only corroborate the biological role of the observed RetS-GacS interactions, but also identify a residue critical for the equilibrium between the RetS-GacS complex and the respective RetS and GacS homodimers. Collectively, our findings suggest that RetS and GacS form a domain-swapped hetero-oligomer during the planktonic growth phase of P. aeruginosa before unknown signals cause its dissociation and a relief of GacS inhibition to promote biofilm formation.
Organizational Affiliation:
Department of Biological Sciences, Virginia Polytechnic Institute and State University, Blacksburg, Virginia, USA.