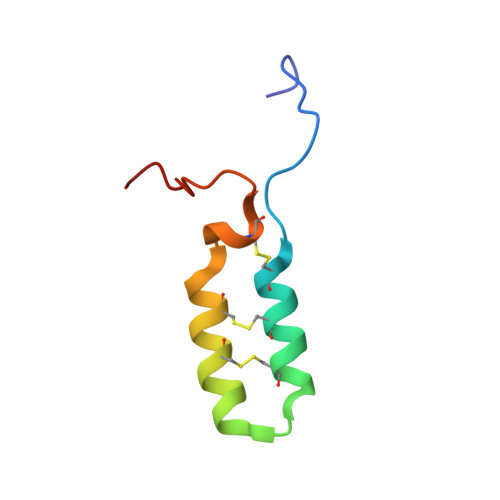
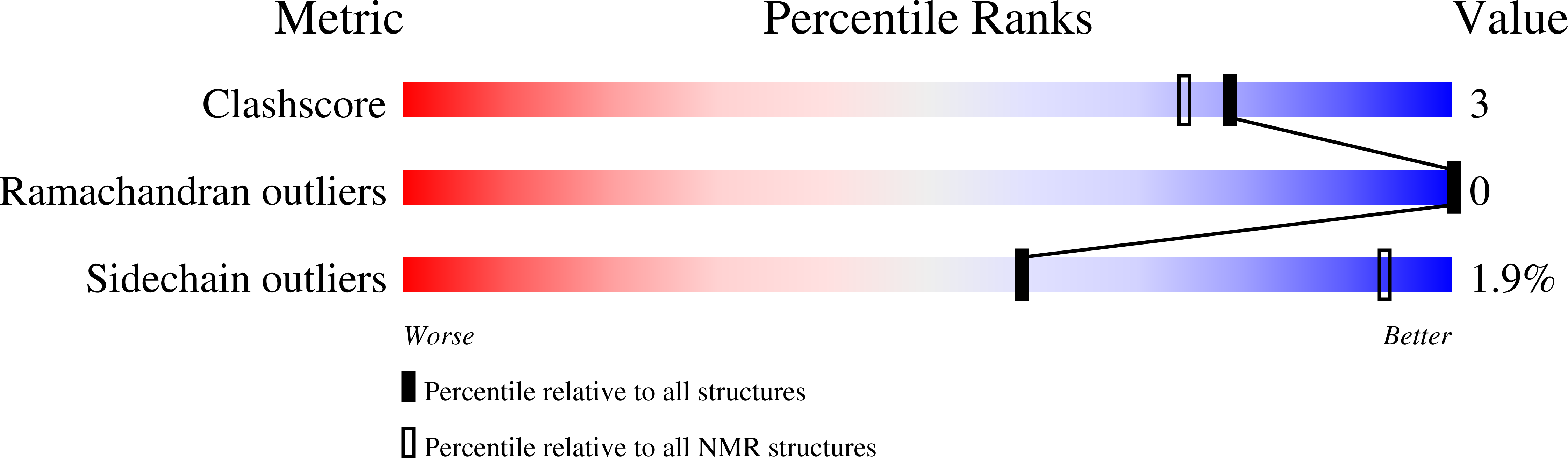Structure, Immunogenicity, and IgE Cross-Reactivity among Walnut and Peanut Vicilin-Buried Peptides.
Foo, A.C.Y., Nesbit, J.B., Gipson, S.A.Y., Cheng, H., Bushel, P., DeRose, E.F., Schein, C.H., Teuber, S.S., Hurlburt, B.K., Maleki, S.J., Mueller, G.A.(2022) J Agric Food Chem 70: 2389-2400
- PubMed: 35139305
- DOI: https://doi.org/10.1021/acs.jafc.1c07225
- Primary Citation of Related Structures:
7LVE, 7LVF, 7LVG, 7LXK - PubMed Abstract:
Vicilin-buried peptides (VBPs) from edible plants are derived from the N-terminal leader sequences (LSs) of seed storage proteins. VBPs are defined by a common α-hairpin fold mediated by conserved CxxxCx (10-14) CxxxC motifs. Here, peanut and walnut VBPs were characterized as potential mediators of both peanut/walnut allergenicity and cross-reactivity despite their low (∼17%) sequence identity. The structures of one peanut (AH1.1) and 3 walnut (JR2.1, JR2.2, JR2.3) VBPs were solved using solution NMR, revealing similar α-hairpin structures stabilized by disulfide bonds with high levels of surface similarity. Peptide microarrays identified several peptide sequences primarily on AH1.1 and JR2.1, which were recognized by peanut-, walnut-, and dual-allergic patient IgE, establishing these peanut and walnut VBPs as potential mediators of allergenicity and cross-reactivity. JR2.2 and JR2.3 displayed extreme resilience against endosomal digestion, potentially hindering epitope generation and likely contributing to their reduced allergic potential.
Organizational Affiliation:
National Institute of Environmental Health Sciences, 111 T.W. Alexander Dr, MD-MR01, Research Triangle Park, North Carolina 27615, United States.