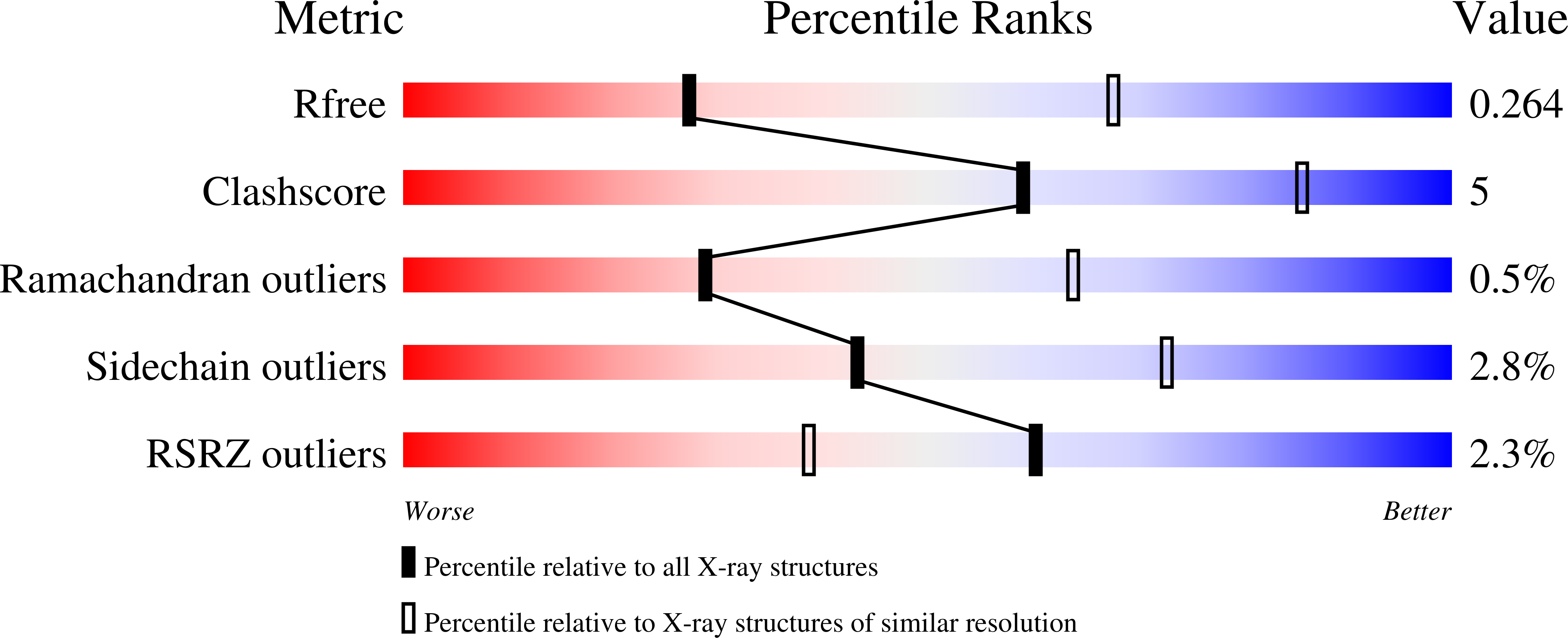
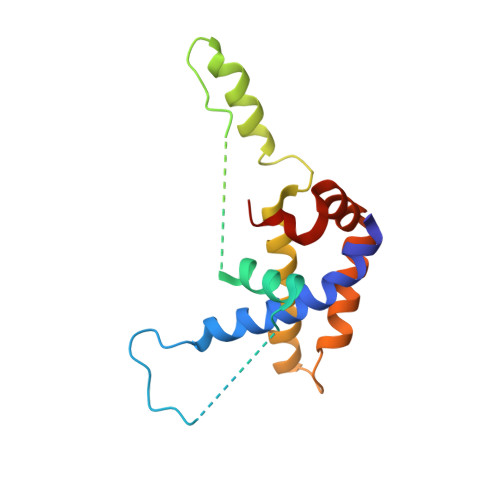
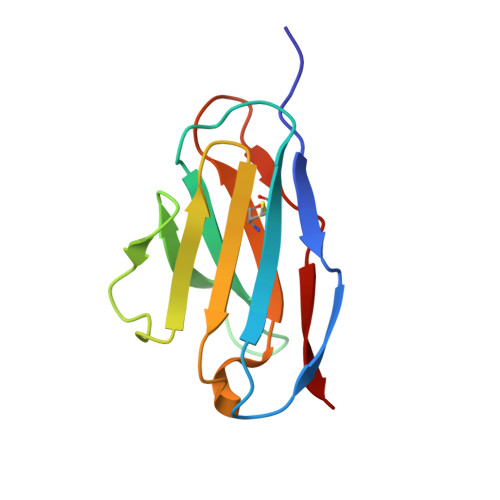
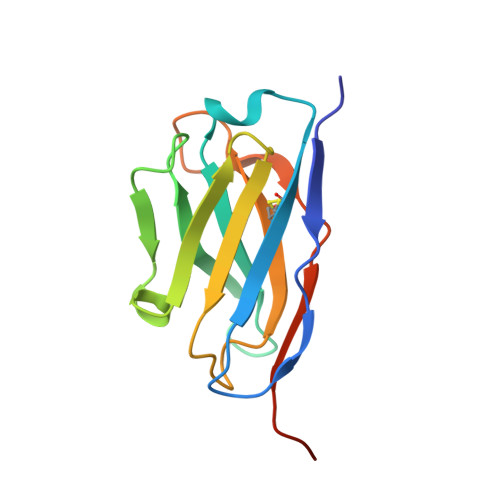
Structure of the BAK-activating antibody 7D10 bound to BAK reveals an unexpected role for the alpha 1-alpha 2 loop in BAK activation.
Robin, A.Y., Miller, M.S., Iyer, S., Shi, M.X., Wardak, A.Z., Lio, D., Smith, N.A., Smith, B.J., Birkinshaw, R.W., Czabotar, P.E., Kluck, R.M., Colman, P.M.(2022) Cell Death Differ 29: 1757-1768
- PubMed: 35279694
- DOI: https://doi.org/10.1038/s41418-022-00961-w
- Primary Citation of Related Structures:
7LK4 - PubMed Abstract:
Pro-apoptotic BAK and BAX are activated by BH3-only proteins to permeabilise the outer mitochondrial membrane. The antibody 7D10 also activates BAK on mitochondria and its epitope has previously been mapped to BAK residues in the loop connecting helices α1 and α2 of BAK. A crystal structure of the complex between the Fv fragment of 7D10 and the BAK mutant L100A suggests a possible mechanism of activation involving the α1-α2 loop residue M60. M60 mutants of BAK have reduced stability and elevated sensitivity to activation by BID, illustrating that M60, through its contacts with residues in helices α1, α5 and α6, is a linchpin stabilising the inert, monomeric structure of BAK. Our data demonstrate that BAK's α1-α2 loop is not a passive covalent connector between secondary structure elements, but a direct restraint on BAK's activation.
Organizational Affiliation:
Structural Biology Division, The Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville, Melbourne, VIC, 3052, Australia.