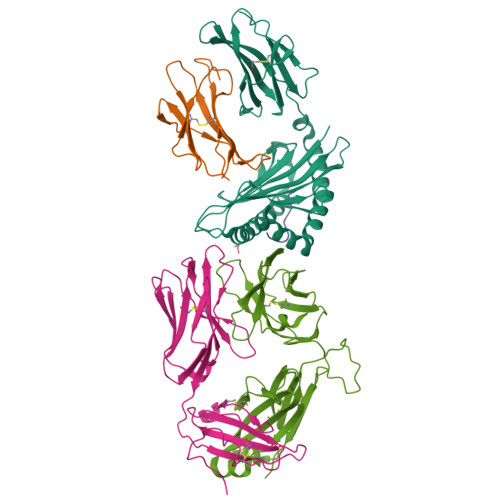
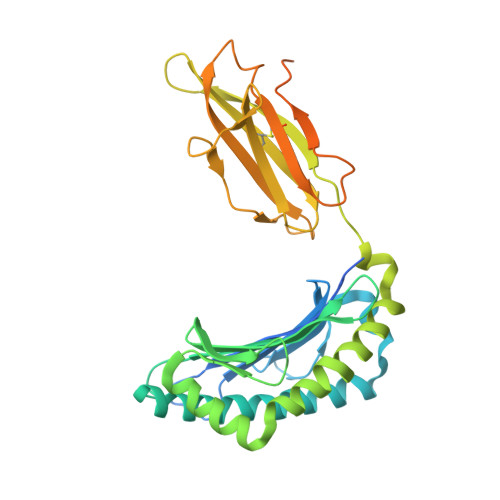
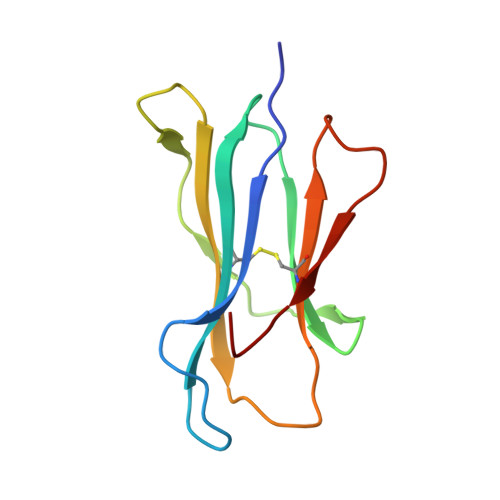
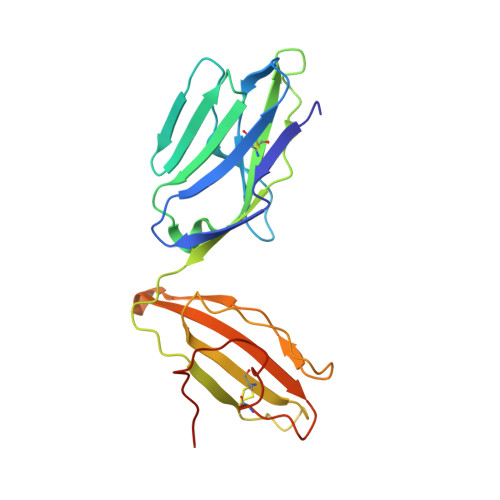
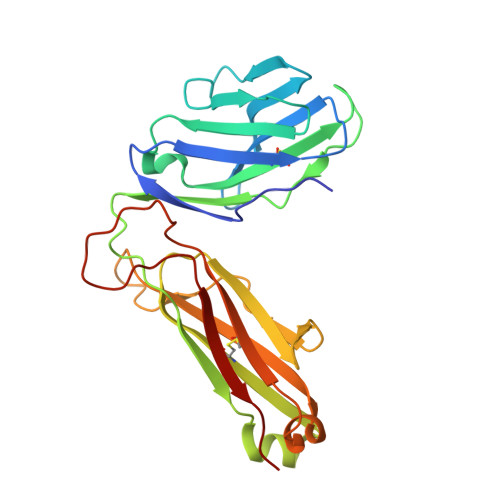
Canonical T cell receptor docking on peptide-MHC is essential for T cell signaling.
Zareie, P., Szeto, C., Farenc, C., Gunasinghe, S.D., Kolawole, E.M., Nguyen, A., Blyth, C., Sng, X.Y.X., Li, J., Jones, C.M., Fulcher, A.J., Jacobs, J.R., Wei, Q., Wojciech, L., Petersen, J., Gascoigne, N.R.J., Evavold, B.D., Gaus, K., Gras, S., Rossjohn, J., La Gruta, N.L.(2021) Science 372
- PubMed: 34083463
- DOI: https://doi.org/10.1126/science.abe9124
- Primary Citation of Related Structures:
7JWI, 7JWJ - PubMed Abstract:
T cell receptor (TCR) recognition of peptide-major histocompatibility complexes (pMHCs) is characterized by a highly conserved docking polarity. Whether this polarity is driven by recognition or signaling constraints remains unclear. Using "reversed-docking" TCRβ-variable (TRBV) 17 + TCRs from the naïve mouse CD8 + T cell repertoire that recognizes the H-2D b -NP 366 epitope, we demonstrate that their inability to support T cell activation and in vivo recruitment is a direct consequence of reversed docking polarity and not TCR-pMHCI binding or clustering characteristics. Canonical TCR-pMHCI docking optimally localizes CD8/Lck to the CD3 complex, which is prevented by reversed TCR-pMHCI polarity. The requirement for canonical docking was circumvented by dissociating Lck from CD8. Thus, the consensus TCR-pMHC docking topology is mandated by T cell signaling constraints.
Organizational Affiliation:
Infection and Immunity Program and Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, Victoria, Australia.