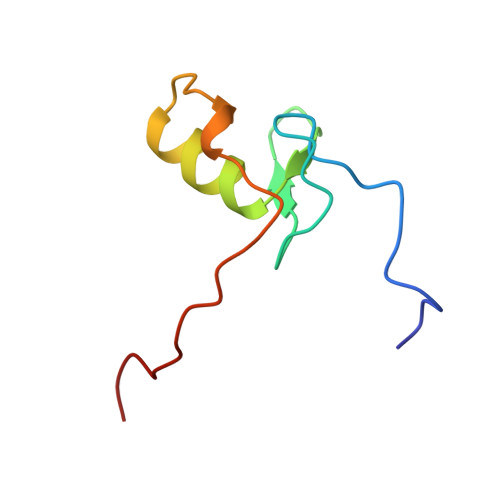
The solution structure of the Leu22-->Val mutant AREA DNA binding domain complexed with a TGATAG core element defines a role for hydrophobic packing in the determination of specificity.
Starich, M.R., Wikstrom, M., Schumacher, S., Arst Jr., H.N., Gronenborn, A.M., Clore, G.M.(1998) J Mol Biol 277: 621-634
- PubMed: 9533884
- DOI: https://doi.org/10.1006/jmbi.1997.1626
- Primary Citation of Related Structures:
6GAT, 7GAT - PubMed Abstract:
The seemingly innocuous leucine-to-valine mutation at position 22 of the AREA DNA binding domain results in dramatic changes in the in vivo expression profile of genes controlled by this GATA transcription factor. This is associated with a preference of the Leu22-->Val mutant for TGATAG sites over (A/C)GATAG sites. Quantitative gel retardation assays confirm this observation and show that the Leu22-->Val mutant AREA DNA binding domain has a approximately 30-fold lower affinity than the wild-type domain for a 13 base-pair oligonucleotide containing the wild-type CGATAG target. To gain insight into the measured affinity data and further explore sequence specificity of the AREA protein, the solution structure of a complex between the Leu22-->Val mutant AREA DNA binding domain and a 13 base-pair oligonucleotide containing its physiologically relevant TGATAG target sequence has been determined by multidimensional nuclear magnetic resonance spectroscopy. Comparison of this structure with that of the wild-type AREA DNA binding domain complexed to its cognate CGATAG target site shows how subtle changes in amino acid side-chain length and hydrophobic packing can affect affinity and specificity for GATA-containing sequences, and how changes in DNA sequence can be compensated for by changes in protein sequence.
Organizational Affiliation:
Laboratory of Chemical Physics, Building 5, National Institute of Diabetes and Digestive and Kidney Diseases National Institutes of Health, Bethesda, MD 20892-0520, USA.