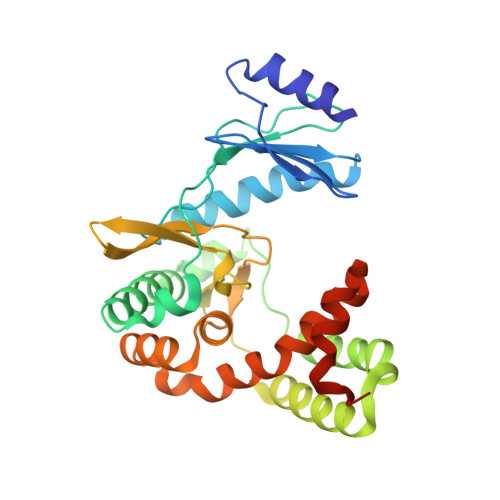
Dual-Mechanism Confers Self-Resistance to the Antituberculosis Antibiotic Capreomycin.
Pan, Y.C., Wang, Y.L., Toh, S.I., Hsu, N.S., Lin, K.H., Xu, Z., Huang, S.C., Wu, T.K., Li, T.L., Chang, C.Y.(2022) ACS Chem Biol 17: 138-146
- PubMed: 34994196
- DOI: https://doi.org/10.1021/acschembio.1c00799
- Primary Citation of Related Structures:
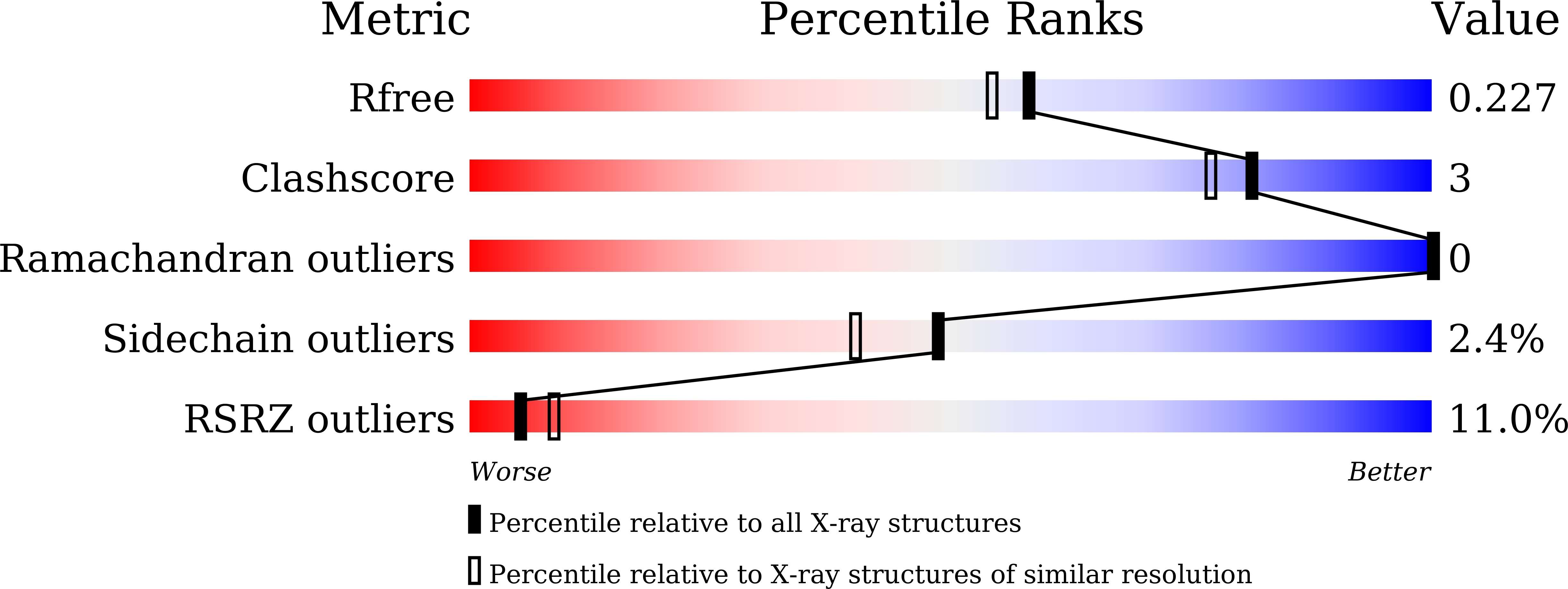
7F0A, 7F0B, 7F0C, 7F0F - PubMed Abstract:
Capreomycin (CMN) is an important second-line antituberculosis antibiotic isolated from Saccharothrix mutabilis subspecies capreolus . The gene cluster for CMN biosynthesis has been identified and sequenced, wherein the cph gene was annotated as a phosphotransferase likely engaging in self-resistance. Previous studies reported that Cph inactivates two CMNs, CMN IA and IIA, by phosphorylation. We, herein, report that (1) Escherichia coli harboring the cph gene becomes resistant to both CMN IIA and IIB, (2) phylogenetic analysis regroups Cph to a new clade in the phosphotransferase protein family, (3) Cph shares a three-dimensional structure akin to the aminoglycoside phosphotransferases with a high binding affinity ( K D ) to both CMN IIA and IIB at micromolar levels, and (4) Cph utilizes either ATP or GTP as a phosphate group donor transferring its γ-phosphate to the hydroxyl group of CMN IIA. Until now, Cph and Vph (viomycin phosphotransferase) are the only two known enzymes inactivating peptide-based antibiotics through phosphorylation. Our biochemical characterization and structural determination conclude that Cph confers the gene-carrying species resistance to CMN by means of either chemical modification or physical sequestration, a naturally manifested belt and braces strategy. These findings add a new chapter into the self-resistance of bioactive natural products, which is often overlooked while designing new bioactive molecules.
Organizational Affiliation:
Department of Biological Science and Technology, National Yang Ming Chiao Tung University, Hsinchu, 30010 Taiwan, R.O.C.