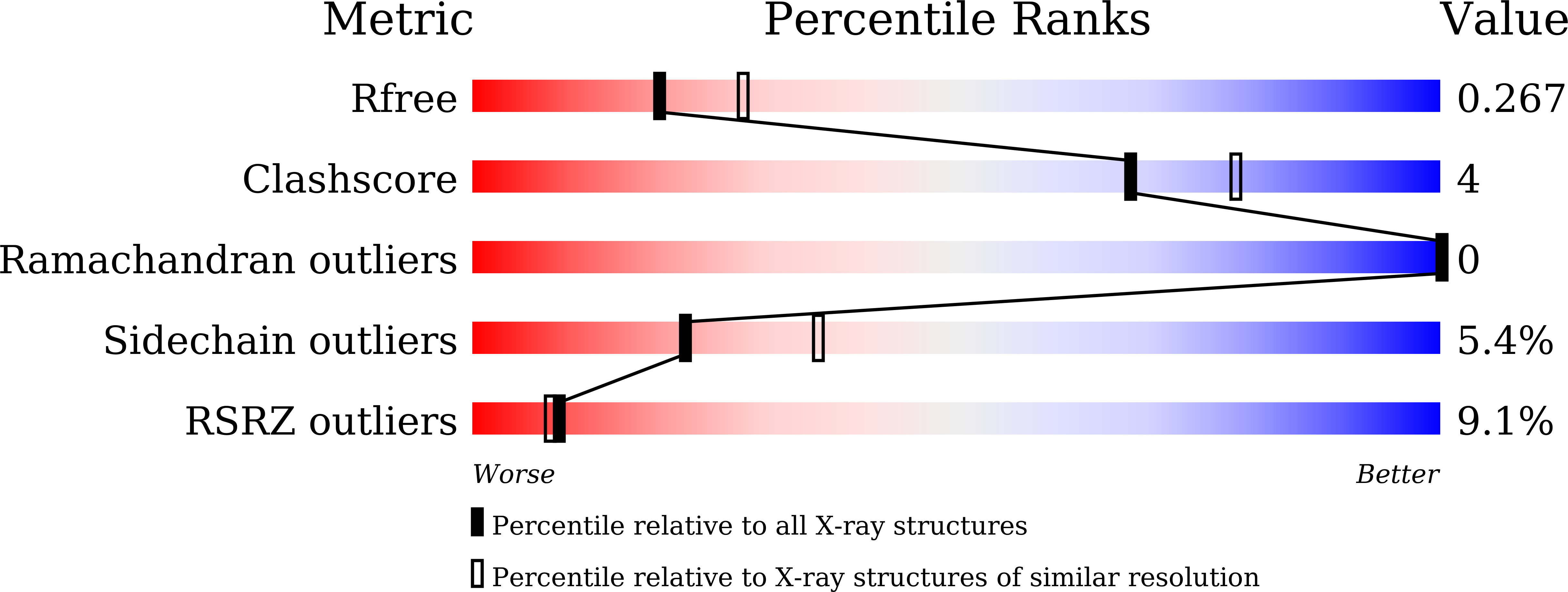
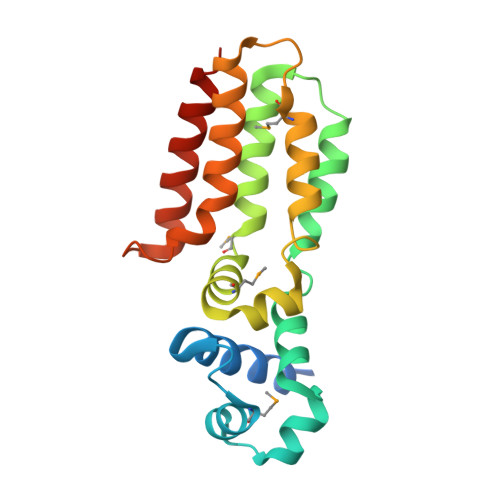
The carbohydrate tail of landomycin A is responsible for its interaction with the repressor protein LanK.
Tsugita, A., Uehara, S., Matsui, T., Yokoyama, T., Ostash, I., Deneka, M., Yalamanchili, S., Bennett, C.S., Tanaka, Y., Ostash, B.(2022) FEBS J 289: 6038-6057
- PubMed: 35429224
- DOI: https://doi.org/10.1111/febs.16460
- Primary Citation of Related Structures:
7EQE, 7EQF - PubMed Abstract:
Landomycin A (LaA) is the largest member of the landomycin group of angucyclic polyketides. Its unusual structure and strong anticancer properties have attracted great interest from chemists and biologists alike. This, in particular, has led to a detailed picture of LaA biosynthesis in Streptomyces cyanogenus S136, the only known LaA producer. LanK is a TetR family repressor protein that limits the export of landomycins from S136 cells until significant amounts of the final product, LaA, have accumulated. Landomycins carrying three or more carbohydrate units in their glycosidic chain are effector molecules for LanK. Yet, the exact mechanism that LanK uses to distinguish the final product, LaA, from intermediate landomycins and sense accumulation of LaA was not known. Here, we report crystal structures of LanK, alone and in complex with LaA, and bioassays of LanK's interaction with synthetic carbohydrate chains of LaA (hexasaccharide) and LaE (trisaccharide). Our data collectively suggest that the carbohydrate moieties are the sole determinants of the interaction of the landomycins with LanK, triggering the latter's dissociation from the lanK-lanJ intergenic region via structure conversion of the helices in the C-terminal ligand-binding domain. Analysis of the available literature suggests that LanK represents an unprecedented type of TetR family repressor that recognises the carbohydrate portion of a natural product, and not an aglycon, as it is the case, for example, with the SimR repressor involved in simocyclinone biosynthesis.
Organizational Affiliation:
Department of Molecular and Chemical Life Sciences, Tohoku University, Sendai, Japan.