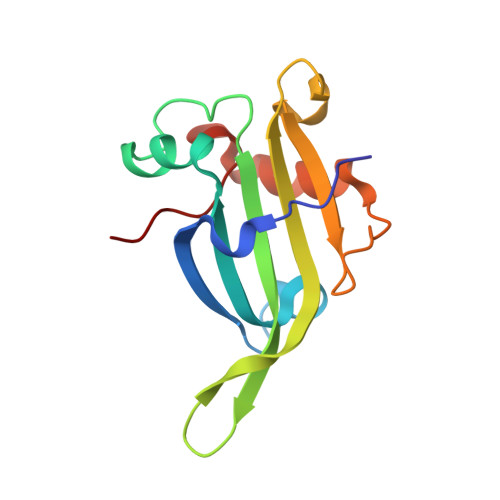
Crystal structure of thermally stable homodimeric cytochrome c'-beta from Thermus thermophilus.
Yoshimi, T., Fujii, S., Oki, H., Igawa, T., Adams, H.R., Ueda, K., Kawahara, K., Ohkubo, T., Hough, M.A., Sambongi, Y.(2022) Acta Crystallogr F Struct Biol Commun 78: 217-225
- PubMed: 35647678
- DOI: https://doi.org/10.1107/S2053230X22005088
- Primary Citation of Related Structures:
7EAD - PubMed Abstract:
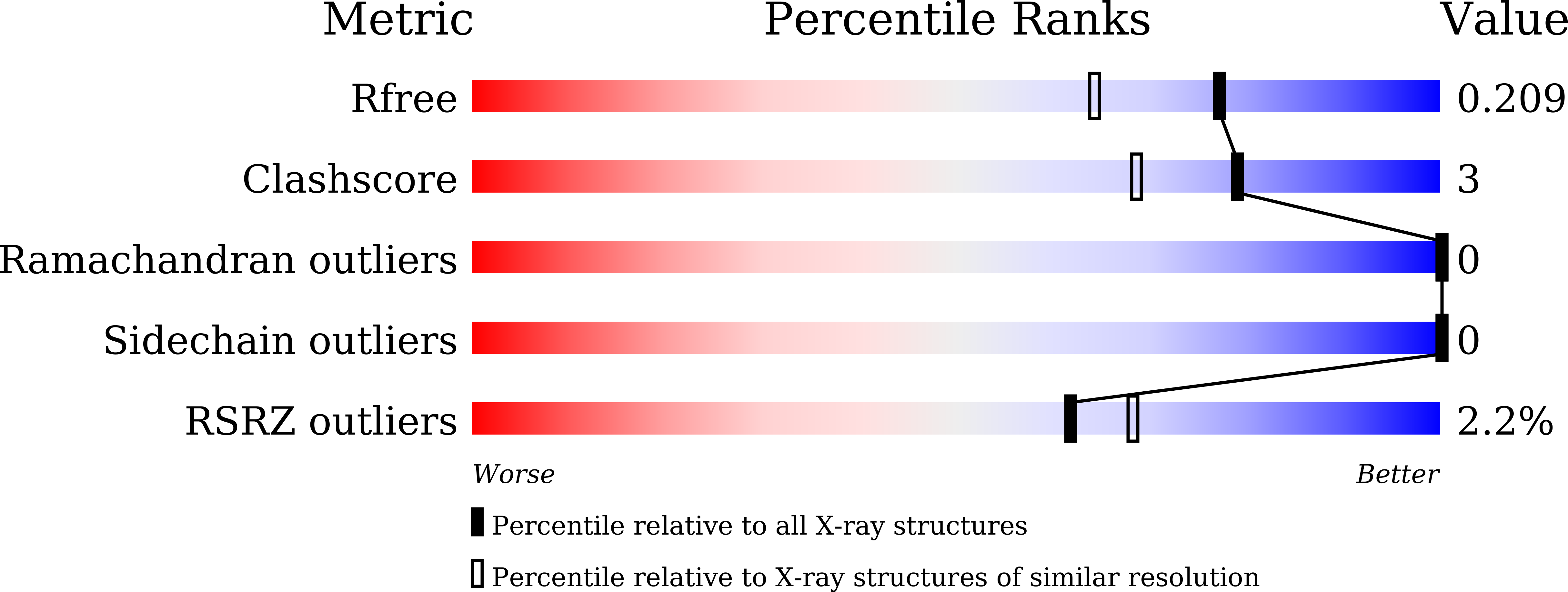
Cytochrome c'-β is a heme protein that belongs to the cytochrome P460 family and consists of homodimeric subunits with a predominantly antiparallel β-sheet fold. Here, the crystal structure of cytochrome c'-β from the thermophilic Thermus thermophilus (TTCP-β) is reported at 1.74 Å resolution. TTCP-β has a typical antiparallel β-sheet fold similar to that of cytochrome c'-β from the moderately thermophilic Methylococcus capsulatus (MCCP-β). The phenylalanine cap structure around the distal side of the heme is also similar in TTCP-β and MCCP-β, indicating that both proteins similarly bind nitric oxide and carbon monoxide, as observed spectroscopically. Notably, TTCP-β exhibits a denaturation temperature of 117°C, which is higher than that of MCCP-β. Mutational analysis reveals that the increased homodimeric interface area of TTCP-β contributes to its high thermal stability. Furthermore, 14 proline residues, which are mostly located in the TTCP-β loop regions, possibly contribute to the rigid loop structure compared with MCCP-β, which has only six proline residues. These findings, together with those from phylogenetic analysis, suggest that the structures of Thermus cytochromes c'-β, including TTCP-β, are optimized for function under the high-temperature conditions in which the source organisms live.
Organizational Affiliation:
Graduate School of Integrated Sciences for Life, Hiroshima University, Higashi-Hiroshima, Japan.