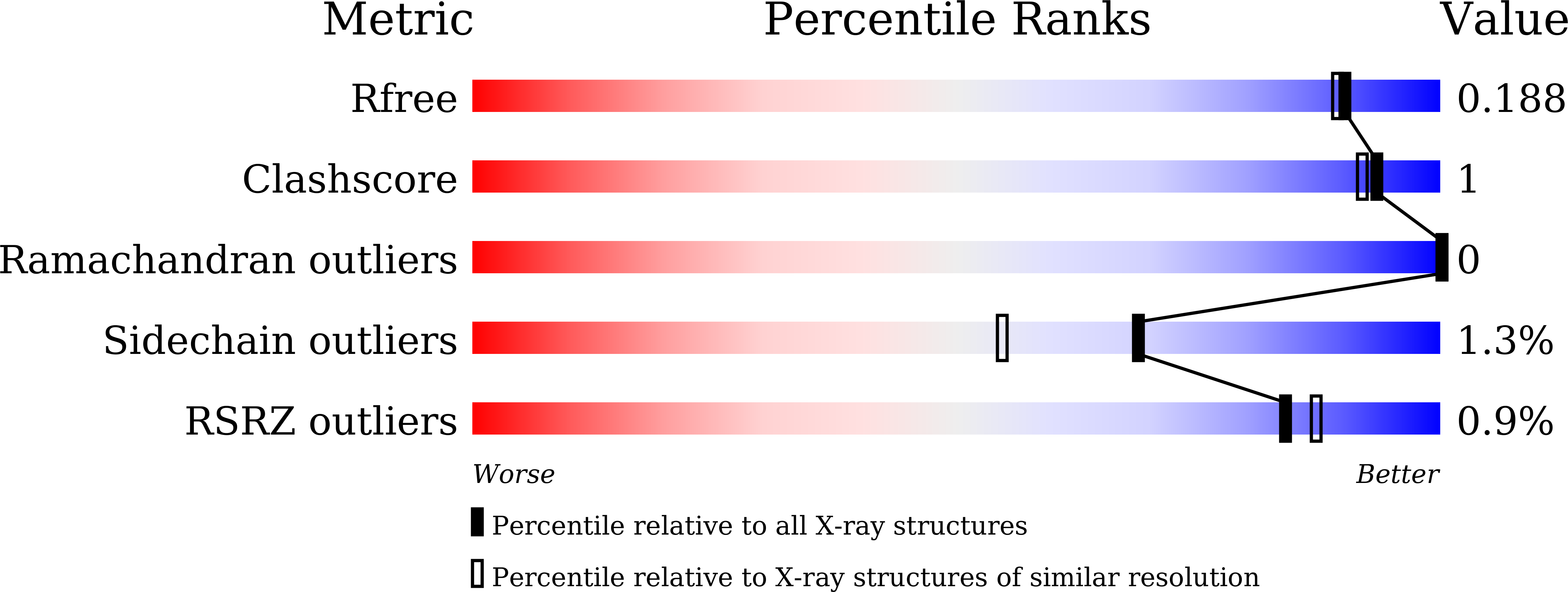
Biochemical and Structural Analysis of a Glucose-Tolerant beta-Glucosidase from the Hemicellulose-Degrading Thermoanaerobacterium saccharolyticum.
Kim, I.J., Bornscheuer, U.T., Nam, K.H.(2022) Molecules 27
- PubMed: 35011521
- DOI: https://doi.org/10.3390/molecules27010290
- Primary Citation of Related Structures:
7E5J - PubMed Abstract:
β-Glucosidases (Bgls) convert cellobiose and other soluble cello-oligomers into glucose and play important roles in fundamental biological processes, providing energy sources in living organisms. Bgls are essential terminal enzymes of cellulose degradation systems and attractive targets for lignocellulose-based biotechnological applications. Characterization of novel Bgls is important for broadening our knowledge of this enzyme class and can provide insights into its further applications. In this study, we report the biochemical and structural analysis of a Bgl from the hemicellulose-degrading thermophilic anaerobe Thermoanaerobacterium saccharolyticum (TsaBgl). TsaBgl exhibited its maximum hydrolase activity on p -nitrophenyl-β-d-glucopyranoside at pH 6.0 and 55 °C. The crystal structure of TsaBgl showed a single (β/α) 8 TIM-barrel fold, and a β8-α14 loop, which is located around the substrate-binding pocket entrance, showing a unique conformation compared with other structurally known Bgls. A Tris molecule inhibited enzyme activity and was bound to the active site of TsaBgl coordinated by the catalytic residues Glu163 (proton donor) and Glu351 (nucleophile). Titration experiments showed that TsaBgl belongs to the glucose-tolerant Bgl family. The gatekeeper site of TsaBgl is similar to those of other glucose-tolerant Bgls, whereas Trp323 and Leu170, which are involved in glucose tolerance, show a unique configuration. Our results therefore improve our knowledge about the Tris-mediated inhibition and glucose tolerance of Bgl family members, which is essential for their industrial application.
Organizational Affiliation:
Department of Biotechnology and Enzyme Catalysis, Institute of Biochemistry, University of Greifswald, 17489 Greifswald, Germany.