Unprecedented Noncanonical Features of the Nonlinear Nonribosomal Peptide Synthetase Assembly Line for WS9326A Biosynthesis.
Kim, M.S., Bae, M., Jung, Y.E., Kim, J.M., Hwang, S., Song, M.C., Ban, Y.H., Bae, E.S., Hong, S., Lee, S.K., Cha, S.S., Oh, D.C., Yoon, Y.J.(2021) Angew Chem Int Ed Engl 60: 19766-19773
- PubMed: 33963654
- DOI: https://doi.org/10.1002/anie.202103872
- Primary Citation of Related Structures:
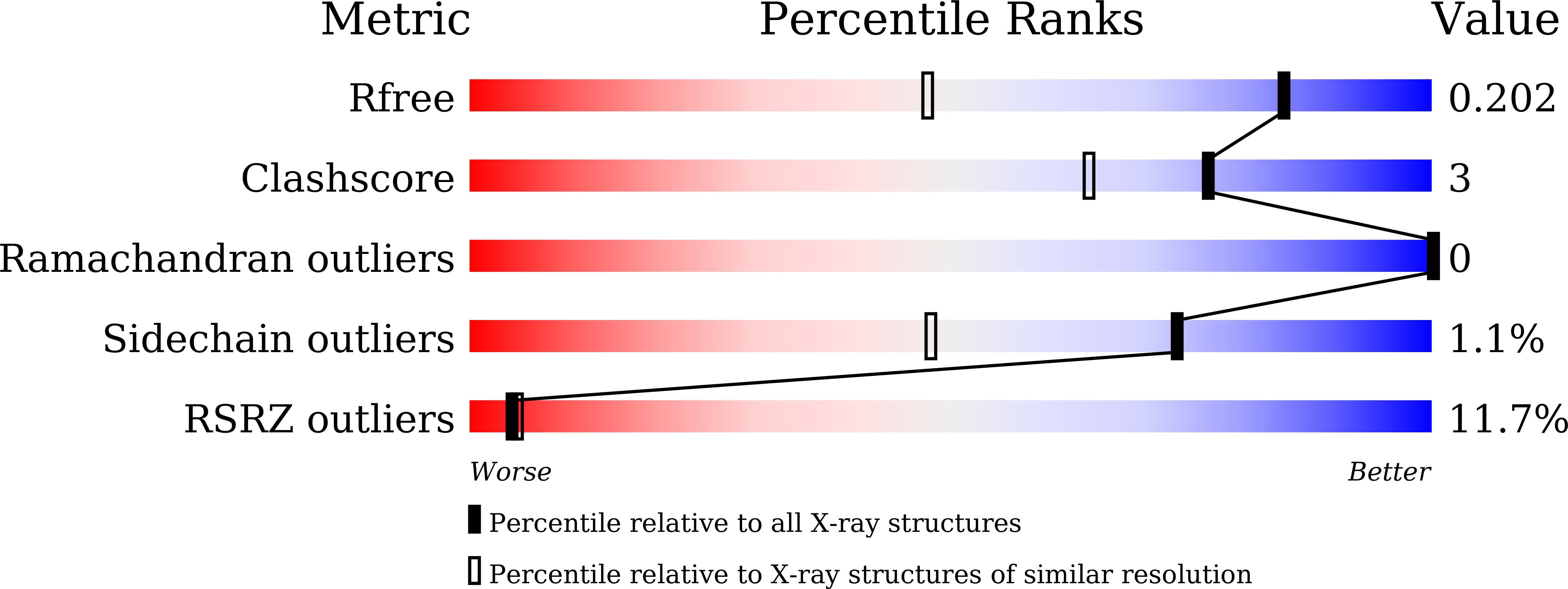
7E3Z - PubMed Abstract:
Systematic inactivation of nonribosomal peptide synthetase (NRPS) domains and translocation of the thioesterase (TE) domain revealed several unprecedented nonlinear NRPS assembly processes during the biosynthesis of the cyclodepsipeptide WS9326A in Streptomyces sp. SNM55. First, two sets of type ΙΙ TE (TEΙΙ)-like enzymes mediate the shuttling of activated amino acids between two sets of stand-alone adenylation (A)-thiolation (T) didomain modules and an "A-less" condensation (C)-T module with distinctive specificities and flexibilities. This was confirmed by the elucidation of the affinities of the A-T didomains for the TEΙΙs and its structure. Second, the C-T didomain module operates iteratively and independently from other modules in the same protein to catalyze two chain elongation cycles. Third, this biosynthetic pathway includes the first example of module skipping, where the interpolated C and T domains are required for chain transfer.
Organizational Affiliation:
Natural Products Research Institute, College of Pharmacy, Seoul National University, 1 Gwanak-ro, Gwanak-gu, Seoul, 08826, Republic of Korea.