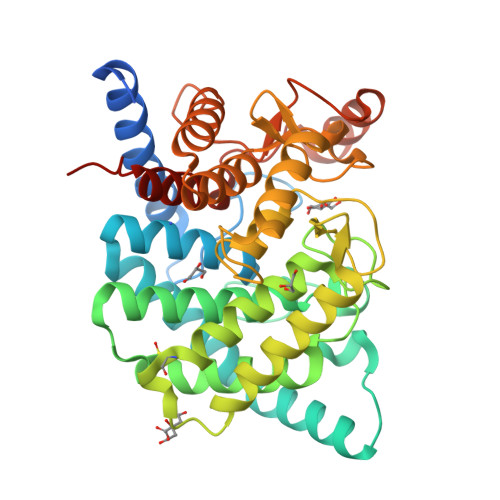
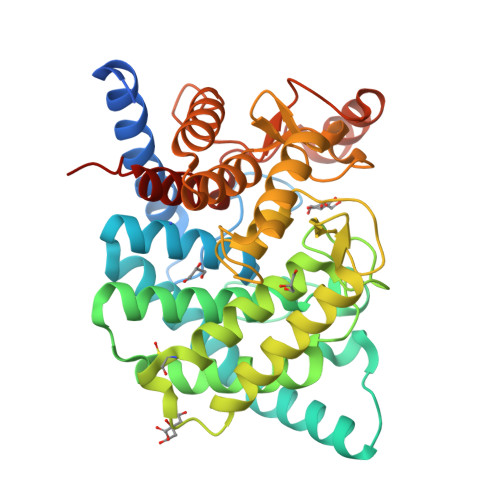
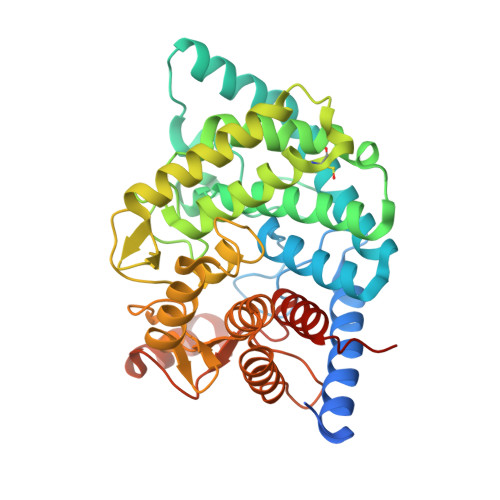
Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Tagami, T., Chen, M., Furunaga, Y., Kikuchi, A., Sadahiro, J., Lang, W., Okuyama, M., Tanaka, Y., Iwasaki, T., Yao, M., Kimura, A.(2022) FEBS J 289: 1118-1134
- PubMed: 34665923
- DOI: https://doi.org/10.1111/febs.16237
- Primary Citation of Related Structures:
5Z3A, 5Z3B, 5Z3C, 5Z3D, 5Z3E, 5Z3F, 7C24, 7C25, 7C26, 7C27 - PubMed Abstract:
Glycoside hydrolase family 15 (GH15) inverting enzymes contain two glutamate residues functioning as a general acid catalyst and a general base catalyst, for isomaltose glucohydrolase (IGHase), Glu178 and Glu335, respectively. Generally, a two-catalytic residue-mediated reaction exhibits a typical bell-shaped pH-activity curve. However, IGHase is found to display atypical non-bell-shaped pH-k cat and pH-k cat /K m profiles, theoretically better-fitted to a three-catalytic residue-associated pH-activity curve. We determined the crystal structure of IGHase by the single-wavelength anomalous dispersion method using sulfur atoms and the cocrystal structure of a catalytic base mutant E335A with isomaltose. Although the activity of E335A was undetectable, the electron density observed in its active site pocket did not correspond to an isomaltose but a glycerol and a β-glucose, cryoprotectant, and hydrolysis product. Our structural and biochemical analyses of several mutant enzymes suggest that Tyr48 acts as a second catalytic base catalyst. Y48F mutant displayed almost equivalent specific activity to a catalytic acid mutant E178A. Tyr48, highly conserved in all GH15 members, is fixed by another Tyr residue in many GH15 enzymes; the latter Tyr is replaced by Phe290 in IGHase. The pH profile of F290Y mutant changed to a bell-shaped curve, suggesting that Phe290 is a key residue distinguishing Tyr48 of IGHase from other GH15 members. Furthermore, F290Y is found to accelerate the condensation of isomaltose from glucose by modifying a hydrogen-bonding network between Tyr290-Tyr48-Glu335. The present study indicates that the atypical Phe290 makes Tyr48 of IGHase unique among GH15 enzymes.
Organizational Affiliation:
Research Faculty of Agriculture, Hokkaido University, Sapporo, Japan.