Computationally designed peptide macrocycle inhibitors of New Delhi metallo-beta-lactamase 1.
Mulligan, V.K., Workman, S., Sun, T., Rettie, S., Li, X., Worrall, L.J., Craven, T.W., King, D.T., Hosseinzadeh, P., Watkins, A.M., Renfrew, P.D., Guffy, S., Labonte, J.W., Moretti, R., Bonneau, R., Strynadka, N.C.J., Baker, D.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 33723038
- DOI: https://doi.org/10.1073/pnas.2012800118
- Primary Citation of Related Structures:
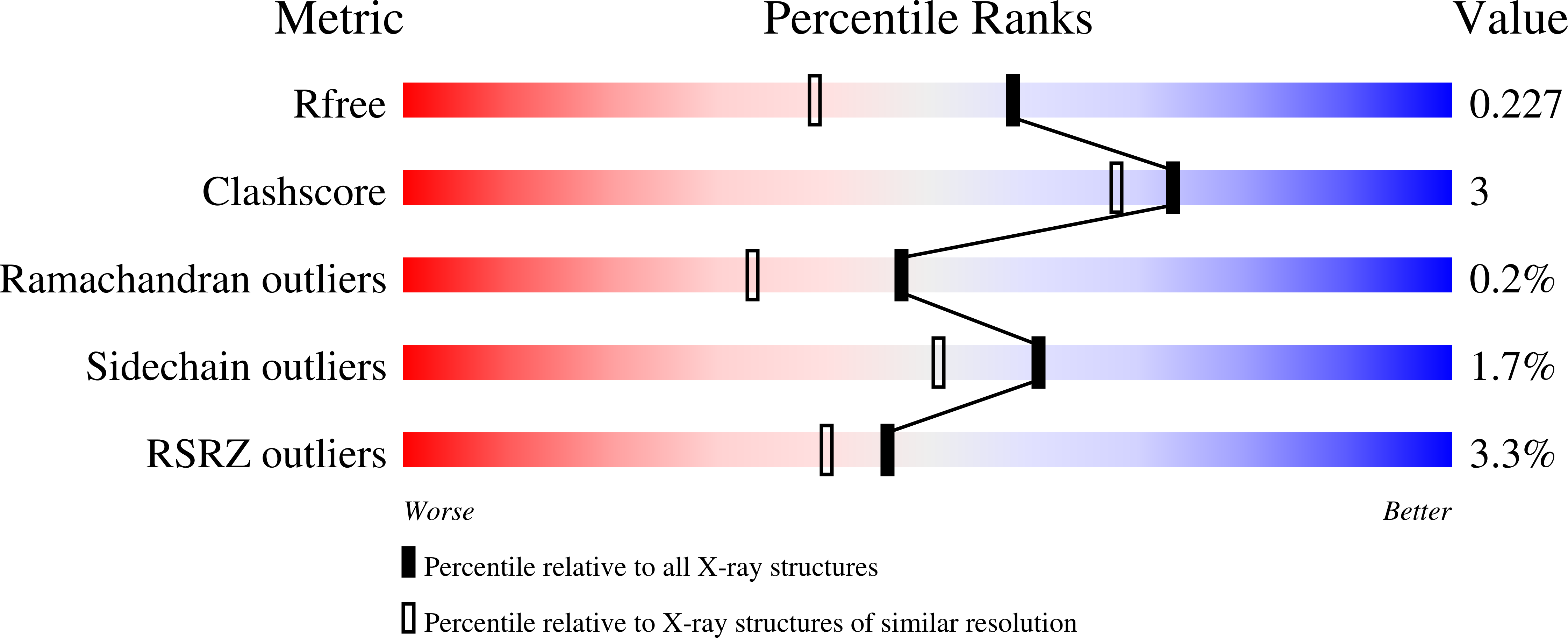
6XBE, 6XBF, 6XCI - PubMed Abstract:
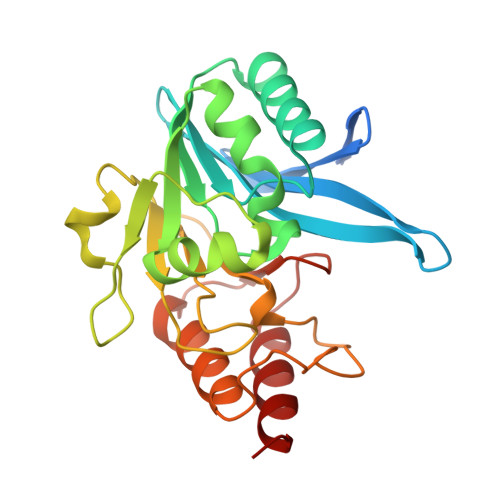
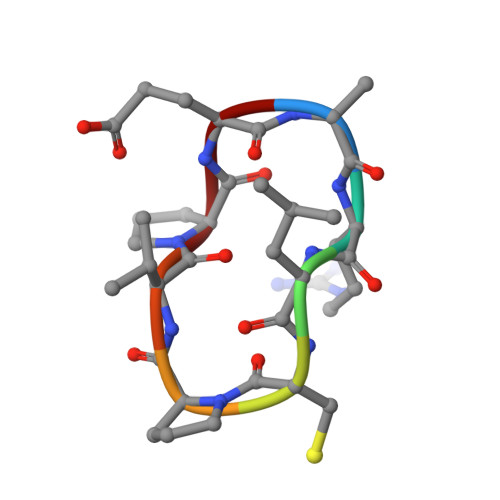
The rise of antibiotic resistance calls for new therapeutics targeting resistance factors such as the New Delhi metallo-β-lactamase 1 (NDM-1), a bacterial enzyme that degrades β-lactam antibiotics. We present structure-guided computational methods for designing peptide macrocycles built from mixtures of l- and d-amino acids that are able to bind to and inhibit targets of therapeutic interest. Our methods explicitly consider the propensity of a peptide to favor a binding-competent conformation, which we found to predict rank order of experimentally observed IC 50 values across seven designed NDM-1- inhibiting peptides. We were able to determine X-ray crystal structures of three of the designed inhibitors in complex with NDM-1, and in all three the conformation of the peptide is very close to the computationally designed model. In two of the three structures, the binding mode with NDM-1 is also very similar to the design model, while in the third, we observed an alternative binding mode likely arising from internal symmetry in the shape of the design combined with flexibility of the target. Although challenges remain in robustly predicting target backbone changes, binding mode, and the effects of mutations on binding affinity, our methods for designing ordered, binding-competent macrocycles should have broad applicability to a wide range of therapeutic targets.
Organizational Affiliation:
Center for Computational Biology, Flatiron Institute, New York, NY 10010; vmulligan@flatironinstitute.org.