Crucial Roles of Two Hydrated Mg2+Ions in Reaction Catalysis of the Pistol Ribozyme.
Teplova, M., Falschlunger, C., Krasheninina, O., Egger, M., Ren, A., Patel, D.J., Micura, R.(2020) Angew Chem Int Ed Engl 59: 2837-2843
- PubMed: 31804735
- DOI: https://doi.org/10.1002/anie.201912522
- Primary Citation of Related Structures:
6UEY, 6UF1, 6UFJ, 6UFK - PubMed Abstract:
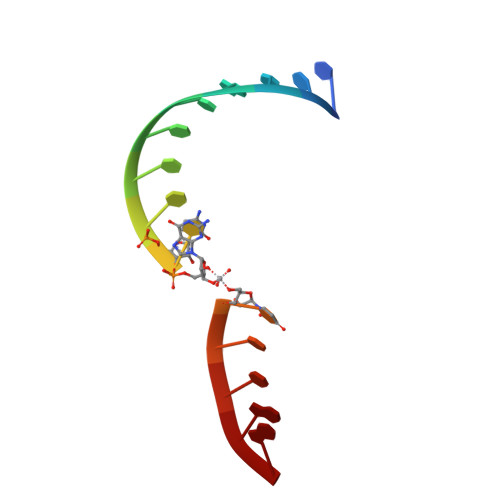
Pistol ribozymes constitute a new class of small self-cleaving RNAs. Crystal structures have been solved, providing three-dimensional snapshots along the reaction coordinate of pistol phosphodiester cleavage, corresponding to the pre-catalytic state, a vanadate mimic of the transition state, and the product. The results led to the proposed underlying chemical mechanism. Importantly, a hydrated Mg 2+ ion remains innersphere-coordinated to N7 of G33 in all three states, and is consistent with its likely role as acid in general acid base catalysis (δ and β catalysis). Strikingly, the new structures shed light on a second hydrated Mg 2+ ion that approaches the scissile phosphate from its binding site in the pre-cleavage state to reach out for water-mediated hydrogen bonding in the cyclophosphate product. The major role of the second Mg 2+ ion appears to be the stabilization of product conformation. This study delivers a mechanistic understanding of ribozyme-catalyzed backbone cleavage.
Organizational Affiliation:
Structural Biology Program, Memorial Sloan-Kettering Cancer Center, New York, New York, 10065, USA.